ПАРАЗИТОЛОГИЯ, 53, 1, 2019
УДК 576.895.121
18S RRNA GENE- AND COX1-GENE-BASED DIVERSITY
OF MONIEZIA SPP. AND AVITELLINA CENTRIPUNCTATA
IN RUMINANTS FROM AL-QADISIYAH PROVINCE, IRAQ
© 2019 M. J. A. Alkaled, Yahia Ismail Khudair,
M. A. A. Al-Fatlawi*
College of Veterinary Medicine, University of Al-Qadisiyah, Al-Diwaniyah, Iraq
*е-mail: monyerr.abd@qu.edu.iq
Submitted 08.06.2018
The present 18S rRNA gene and Cox1 gene-based work was proposed and performed to detect and
study the history of evolution of tapeworms that could be recognized through the use of molecular
techniques. For this investigation, 14 tapeworms were recovered from 50 intestinal-based content
samples of 50 animals (sheep, goat, cattle, and buffalo). The collected adult tapeworms were first
identified morphologically showing 6 Moniezia expansa and 7 Moniezia benedeni. Then, DNA from
14 worms (9 for the 18S rRNA gene and 5 for the Cox1 gene) was employed for polymerase chain
reaction (PCR) and partial gene sequencing techniques. The results of the PCR recorded amplifications
of all the sequences used for both genes. The sequencing indicated, via the use of 18S rRNA gene,
5 Moniezia benedeni, 3 Moniezia expansa, and 1 Avitellina centripunctata. Moreover, it resulted, via
the utilization of Cox1 gene, 3 Moniezia expansa and 2 Moniezia benedeni. The isolates stood in close
matching with global isolates. The current results provide important criteria for the reliable use of the
techniques utilized in this study.
Keywords: 18S rRNA, Avitellina, Cox1, Moniezia, PCR, sequencing.
DOI: 10.1134/S0031184719010034
ВЫЯВЛЕНИЕ РАЗНООБРАЗИЯ ЦЕСТОД MONIEZIA SPP. И AVITELLINA
CENTRIPUNCTATA ИЗ ЖВАЧНЫХ ЖИВОТНЫХ В ПРОВИНЦИИ АЛЬ-
КАДИСИЯ В ИРАКЕ С ИСПОЛЬЗОВАНИЕМ ГЕНОВ 18S РРНК И COX1
© 2019 г. M. J. A. Alkaled, Yahia Ismail Khudair, M. A. A. Al-Fatlawi
Колледж ветеринарной медицины, Университет Аль-Кадисии, Эд-Дивания, Ирак
е-mail: monyerr.abd@qu.edu.iq
Поступила 08.06.2018 г.
Молекулярно-генетическое исследование по генам 18S рРНК A и Cox1 предпринято для ре-
конструкции эволюции цестод жвачных животных. Исследовано 14 экз. цестод из кишечников
31
50 животных (овцы, козы, коровы, буйволы). Вначале половозрелые цестоды были идентифи-
цированы морфологически: 6 экз. Moniezia expansa и 7 экз. Moniezia benedeni. Затем ДНК 14
червей (9 для анализа 18SрРНК и 5 для Cox1) были использованы для ПЦР и секвенирования
участков генов. По результатам секвенирования участка гена 18S рРНК выявлены 5 экз. Moniezia
benedeni, 3 экз. Moniezia expansa и 1 экз. Avitellina centripunctata. С помощью анализа гена Cox1
выявлены 3 экз. Moniezia expansa и 2 экз. Moniezia benedeni. Обнаружено близкое совпадение с
последовательностями нуклеотидов генов в глобальном банке данных. Полученные результаты
помогли установить критерии надежного использования молекулярных методик идентификации
цестод.
Ключевые слова: 18S rRNA, Avitellina, Cox1, Moniezia, ПЦР, секвенирование.
Moniezia spp are tapeworms that infest various ruminants such as sheep, goat, cattle, and
buffalos. These worms induce some problems to the health of animals plus to industries
leading to financial losses via the increase cost provided for the treatment and veterinary
services (Kouam et al., 2018). Two species are well-known for their infestation effectiveness
on animal industries; Moniezia benedeni and Moniezia expansa. For the morphological
recognition of these tapeworms, characteristics features are presented on them such as scolex,
neck and strobila. The scolex and neck are measured to be small in sizes with noticed-long
chain of strobili. These interesting worms belong to the family of Anoplocephalidae and the
order of Cyclophyllidea. Extra differences of Moniezia genus that are characterized by the
presence of clear anterior, posterior, mature, and gravid segments of these worms. Another
characteristic feature is that each proglottid has repeated-sexual parts. Moniezia spp. needs
mites as an intermediate host for the lifecycle to be completed via the presence of grass
feeding by the affected animals (Denegri et al., 1998; Aboma et al., 2015; Ohtori et al.,
2015). Avitellina centripunctata is cestode that infests sheep, goats, cattle, and wide range of
ruminants (Yildiz, 2007). The strobili of these worms are characterized by the presence of
thousands of wide-very short proglottids. The lifecycle needs certain species of arthropods,
oribatid mites and barklice, to be completed (Woodland, 1935; Yildiz, 2007). The present
18S rRNA gene and Cox1 gene based work was proposed and performed to detect and study
the history of evolution of tapeworms that could be recognized through the use of molecular
techniques that rely on 18S rRNA and Cox1 genes. For this investigation, 14 tapeworms were
recovered from 50 intestinal-based content samples of 50 animals (sheep, goat, cattle, and
buffalo). PCR and Partial gene sequencing were employed for this present work to fulfill
the goal of the study.
MATERIALS AND METHODS
Sampling
For this work, 50 intestinal-based content samples were collected from 50 animals (sheep,
goat, cattle, and buffalo). The collection work continued from August, 2016 to January, 2017.
The locations of sampling were from different slaughter houses in Al-Qadisiyah province,
Iraq. The tapeworms were placed separately in clean containers that contained PBS. At the
Lab in the college of veterinary medicine, University of Al-Qadisiyah, adult worms were
placed in containers that had 70 % ethanol and stored at -20°C until future work, starting
with DNA extraction. Some of these tapeworms were subjected to morphological-based
identification.
32
DNA extraction
A piece of the worm, 200mg, was first rinsed thoroughly with water to get rid of ethanol.
KAPA Express Extract Kit (Cape Town, South Africa) or (Roche, Mannheim, Germany) was
used to fulfill the extraction process relying on the protocol provided by the manufacturer.
A NanoDrop was utilized to measure the resulted DNA for quality and quantity.
PCR and sequencing techniques
The technique followed the use of specific primers that were designed using
PrimerQuest Tool (Integrated DNA Technologies, Inc., Belgium) and NCBI-based
websites. The primers of the 18S rRNA gene were F: ACGGTGAAACCGCGAATGG and
R: GACATGACATGCAGTAGCAGTG that amplify a specific region in this gene at
841 bp. Moreover for the Cox1 gene, the primers were F: TGTTGAGTATGTGGTTTGGTGC
and R: AACTACCCACCATACCACAGGATC that amplify a region at 684 bp. DNA from
14 worms (9 for the 18S rRNA gene and 5 for the Cox1 gene) was employed for the PCR. The
kit that was used for the preparation of the mastermix was ordered from (Bionear, Korea).
The instructions of the kit were followed for this purpose. Regarding the conditions of the
thermocycler used were 95°C for 5 min as for the primary denaturation, 35 cycles of (main
denaturation at 94°C for 1 min, for the 18S rRNA and Cox1 55°C and 53°C respectively for
the annealing, for the 18S rRNA and Cox1 55°C for 1 min and 72°C for 2.5 min respectively),
and 72°C for 5 min regarding the finishing extension. the PCR products in the 1.25 % agarose
gels were electrophoresed and visualized via the use of ethidium-bromide-based illumination
screened by a UV imager. All PCR products were sent out for partial-gene sequencing using
Sanger sequencing method (Macrohen Company, Korea). The phylogenetic tree was built up
using maximum likelihood via the use of MEGA 7.0 (Saitou, Nei, 1987; Tamura et al., 2013).
RESULTS
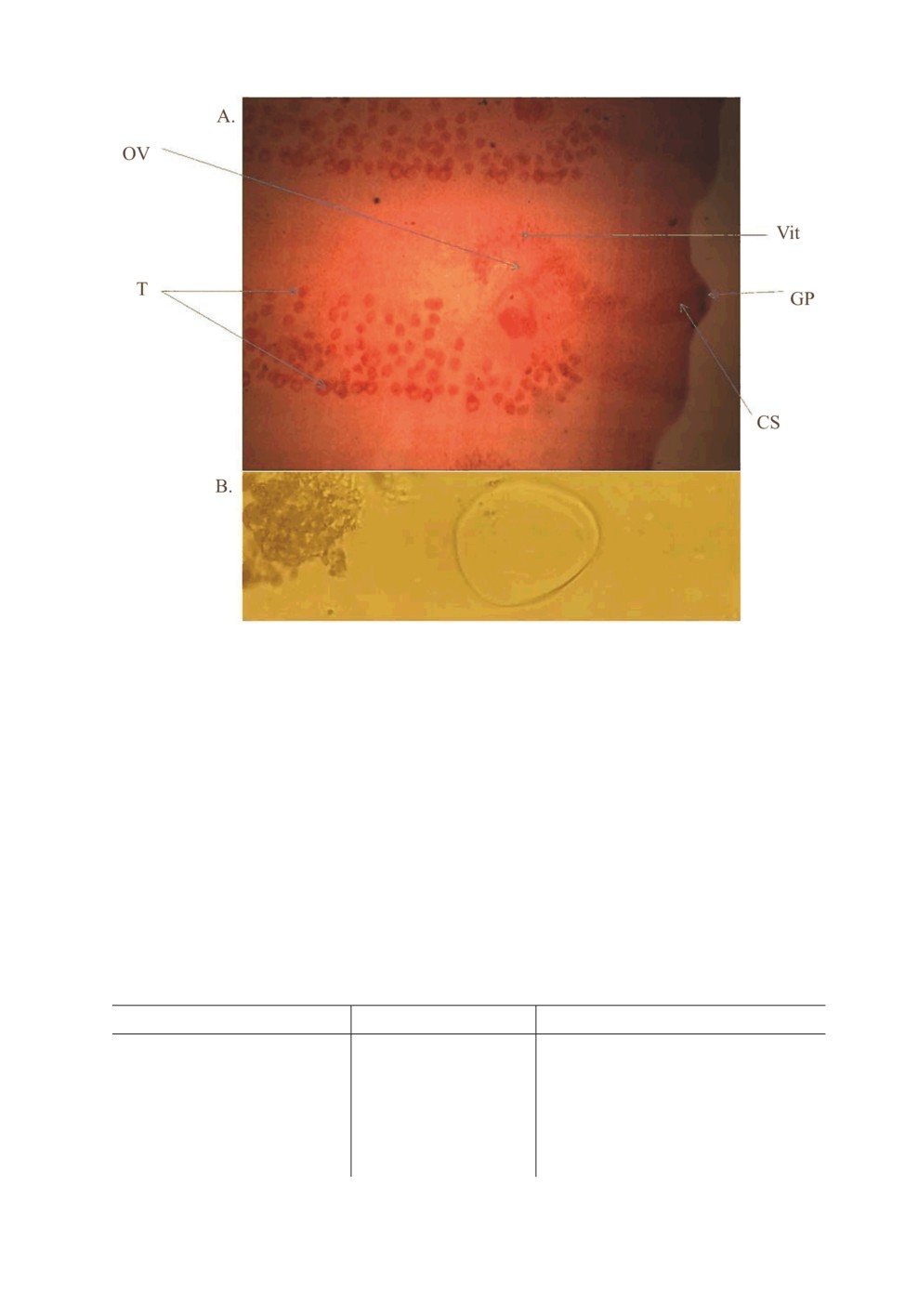
According to the morphological characters, worms belonging to the genus Moniezia were
recognized. The generic characters were as follows: genital pore, clear cirrus sac, vitelline
gland, recognizable testes, and inter-proglottid-based glands. For the species identification,
adult worm and egg features were used to separate between M. expansa and M. benedeni
that were recovered in this study showing 6 Moniezia expansa and 7 Moniezia benedeni
(figs. 1, 2).The results of the PCR recorded amplifications of all the sequences used for both
genes (figs. 3, 4).
The sequencing indicated, via the use of 18S rRNA gene, 5 Moniezia benedeni, 3 Moniezia
expansa, and 1 Avitellina centripunctata. Moreover, it resulted, via the utilization of
Cox1 gene, 3 Moniezia expansa and 2 Moniezia benedeni. The current isolates, MH173843.1,
MH173844.1, MH173845.1, MH173846.1, MH173847.1, MH173848.1, MH201211.1,
MH201212.1 MH201213.1, and MH201214.1 stood in close matching with global isolates
(figs. 5, 6). Tables 1 and 2 provide statistics about the isolates regarding their sampled animals.
DISCUSSION
Moniezia spp. are tapeworms that infest various ruminants such as sheep, goat, cattle,
and buffalos. These worms induce some problems to the health of animals plus to
industries leading to financial losses via the increase costs provided for the treatment and
veterinary services (Diop et al., 2015a; Kouam et al., 2018). Two species are well-known
33
Fig. 1. A. Moniezia expansa. IPG = inter-proglottidal glands, T = Testes, Vit = Vittline gland,
CS = Cirrus sac, GP = Genital pore, OV = Ovary. (X10) (We can see the IPG because they reach
to the margin of segments. B. Moniezia expansa egg (Quadral form). (X10).
Table 1. Shows statistics about the isolates regarding their sampled animals via the use of 18S rRNA
gene
Animal
ID
Cestode species
Cattle
DIQ2
M. benedeni
Cattle
DIQ3
M. benedeni
Cattle
DIQ4
M. benedeni
Sheep
DIQ5
M. benedeni
Sheep
DIQ9
Avitellina centripunctata
Buffalo
DIQ6
M. benedeni
Buffalo
DIQ10
M. expansa
Goat
DIQ11
M. expansa
Goat
DIQ12
M. benedeni
34
Fig. 2. A. Moniezia benedeni, T = Testes, Vit = Vittline gland, CS = Cirrus sac, GP = Genital pore,
OV = Ovary. (X10) (The inter-proglottidal glands do not reach to the margin of segments).
B. Moniezia benedeni egg (Triangle). (X40).
for their infestation effectiveness on animal industries. The present study was successful in
determining the presence of Moniezia spp. via the morphological identification. This step
is important in the diagnosis any parasites especially tapeworms that have their external
characteristic features. The use of the PCR technique confirm that worms are identical to
the genus Moniezia, thus confirming the previous studies (Mehlhorn, 2016). These results
agree with (Nguyen et al., 2012) who detected tapeworms via the use of a PCR method. The
18S rRNA gene sequencing analysis revealed 5 Moniezia benedeni, 3 Moniezia expansa,
and 1 Avitellina centripunctata. This technique is proved to be reliable in identifying these
tapeworms, and it is useful in detecting more species than that in the other method, that
Table 2. Shows statistics about the isolates regarding their sampled animals via the use of Cox1 gene
Animal
ID
Cestode species
Cattle
DIQ14
M. benedeni
Cattle
DIQ15
M. benedeni
Sheep
DIQ13
M. expansa
Sheep
DIQ12
M. expansa
Buffalo
DIQ11
M. expansa
35
Fig. 3. Image of the PCR products via gel-based electrophoresis of the 18S rRNA gene.
Positive lanes are 1-8. Ladder is M lane.
Fig. 4. Image of the PCR products via gel-based electrophoresis of the Cox1 gene.
Positive lanes are 1, 3, 4, and 5. Ladder is M lane.
36
Fig. 5. Phylogenetic tree based on the 18S rRNA gene partial sequencing.
utilized Cox1 as a target gene. However, it was consistent, via the utilization of Cox1 gene,
in detecting 3 Moniezia expansa and 2 Moniezia benedeni. The current isolates were placed
in close matching with global isolates on the phylogenetic tree. However, our isolates have
distinct mutations that may occur in the past and let them to branch out from the global
isolates (Yan et al., 2013; Ohtori et al., 2015). The results of the phylogenetic tree may
indicate that these isolates of the sampled animals were from different ancestors (Diop
et al., 2015b; Ohtori et al., 2015; Guo, 2016). In a recent published work by (Haukisalmi
et al., 2018), 6 Moniezia species were identified in 2 clades. Although their results showed
M. expansa and M. benedeni in Clade1 of their phylogenetic tree, some of the 6 species were
37
Fig. 6. Phylogenetic tree based on the Cox1 gene partial sequencing.
organized in the Clade2. Our results agree to some extent with this work especially when
look at our tree that placed these both species in different clades. These variations could have
been initiated because the geographical differences between Iraq and Finland and Alaska. In
Iraq, the disease control systems may not be as good as the ones in those places, and this may
introduce certain impacts on generating modifications in the nucleotide sequences leading to
the appearance of new strains. The current results provide important criteria for the reliable
use of the techniques utilized in this study.
REFERENCES
Aboma R., Nesibu A., Birhanu H., Yisehak T., Teshale S. 2015. Internal and external parasites of camels (Camelus
dromedarius) slaughtered at Addis Ababa Abattoir, Ethiopia. Journal of Veterinary Medicine and Animal
Health 7 (2): 57-63.
Denegri G., Bernadina W., Perez-Serrano J., Rodriguez-Caabeiro F. 1998. Anoplocephalid cestodes of veterinary
and medical significance: a review. Folia Parasitologica (Praha) 45 (1): 1-8.
Diop G., Yanagida T., Hailemariam Z., et al. 2015a. Genetic characterization of Moniezia species in Senegal and
Ethiopia. Parasitology International 64 (5): 256-260.
Diop G., Yanagida T., Hailemariam Z., et al. 2015b. Genetic characterization of Moniezia species in Senegal and
Ethiopia. Parasitology International 64 (5): 256-260.
Guo A. 2016. Moniezia benedeni and Moniezia expansa are distinct cestode species based on complete mitochondrial
genomes. Acta Tropica 166: 287-292.
Haukisalmi V. et al. 2018. Molecular taxonomy and subgeneric classification of tapeworms of the genus Moniezia
Blanchard, 1891 (Cestoda, Anoplocephalidae) in northern cervids (Alces and Rangifer). Parasitology
International 67 (2): 218-224.
38
Kouam M.K., Meningue R., Fon D.E. 2018. Parasitic causes of organ condemnation in cattle slaughtered in Fako
abattoirs, South-West region of Cameroon, and estimate of financial losses. Journal of Helminthology 2018:
1-5.
Mehlhorn H. (Ed.) 2016. Encyclopedia of Parasitology. 4th edition. Springer, 3096 pp.
Nguyen T.D., Le Q.D., Huynh V.V., Nguyen S.T., Nguyen T.V., Vu-Khac H. 2012. The development of
PCR methodology for the identification of species of the tapeworm Moniezia from cattle, goats and sheep
in central Vietnam. Journal of Helminthology 86 (04): 426-429.
Ohtori M., Aoki M., Itagaki T. 2015. Sequence differences in the internal transcribed spacer 1 and 5.8S ribosomal
RNA among three Moniezia species isolated from ruminants in Japan. Journal of Veterinary Medical Science
77 (1): 105-107.
Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees.
Molecular Biology and Evolution 4 (4): 406-425.
Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis
version 6.0. Molecular Biology and Evolution 30 (12): 2725-2729.
Woodland W.N.F. 1935. A New Species of Avitellinine Tapeworm, Avitellina Sandgroundi , from Hippotragus
Equinus. Annals of Tropical Medicine and Parasitology 29 (2): 185-189.
Yan H., Bo X., Liu Y., et al. 2013. Differential diagnosis of Moniezia benedeni and M. expansa (Anoplocephalidae)
by PCR using markers in small ribosomal DNA (18S rDNA). Acta Veterinaria Hungarica 61 (4): 463-472.
Yildiz K. 2007. The scanning electron microscopic examination of Avitellina centripunctata and Thysaniezia ovilla.
Turkiye parazitolojii Derg 31 (4): 292-295.
39