Микология и фитопатология, 2019, T. 53, № 2, стр. 108-114
Studies on the mycobiota of blighted Solanum dulcamara leaves
L. Yu. Kokaeva 1, 2, Yu. I. Berezov 1, S. V. Zhevora 2, P. N. Balabko 1, E. M. Chudinova 3, E. Yu. Voronina 1, S. N. Elansky 1, 3, *
1 Lomonosov Moscow State University
119991 Moscow, Russia
2 All-Russian Lorh Research Institute of Potato Farming
143050 Moscow Region, Kraskovo, Russia
3 Peoples Friendship University of Russia
117198 Moscow, Russia
* E-mail: snelansky@gmail.com
Поступила в редакцию 29.03.2018
После доработки 30.03.2018
Принята к публикации 29.05.2018
Аннотация
Mycobiota of affected bittersweet nightshade (Solanum dulcamara) leaves has been studied. The leaves were collected on the territory of Lomonosov Moscow State University (Vorobyovy gory, Moscow). Sampled leaves were instantly frozen at –75°С for DNA extraction or placed immediately into moist chambers for isolation of fungal cultures. DNA was extracted from the whole leaf. Ribosomal DNA (including ITS1 and ITS2 regions) gene banks were constructed in Escherichia coli by cloning PCR products generated with primer pairs ITS1f and ITS4. Fragments from the cloned inserts were sequenced and compared to known rDNA sequences. Sixteen fungal species were revealed by cloning and isolation of fungal cultures: Alternaria alternata, Aureobasidium pullulans, Boeremia exigua (= Phoma exigua), Botrytis cinerea, Cladosporium cladosporioides, C. herbarum, C. tenuissimum, Colletotrichum acutatum, Colletotrichum gloeosporioides, Coniothyrium fuckelii, Fusarium oxysporum, Phoma herbarum, Phytophthora infestans, Thanatephorus cucumeris, Thielavia basicola. Four taxa were identified at the level of genera (Cryptococcus sp., Mycosphaerella sp., Phialophora sp., Phoma sp.). All collected leaf samples contained from 5 to 8 different fungal taxa. Most of the found species were known pathogens of solanaceous plants. Some species could produce toxins or cause human allergies.
Wild Solanum species inhabit urban waste lands and roadsides and may be found close to potato and tomato plantations. Sometimes Solanum species are used as ornamentals for urban and suburban gardening; however, they can act as alternate hosts for pathogens affecting solanaceous crops (potato, tomato, eggplant, and other). Bittersweet nightshade (Solanum dulcamara L.), which is a perennial liana, belongs to the most wide-spread wild Solanaceae species of Central Russia. S. dulcamara plants may provide a natural late blight depositary (Cooke et al., 2002; Flier et al., 2003).
S. dulcamara plants with late blight lesions have already been found on the territory of the Moscow State University. The pathogenicity and spore production of Phytophthora infestans (Mont.) de Bary isolates collected from this plant were determined using artificially infected potato and tomato leaves (Elansky et al., 2015). Bittersweet nightshade could also serve as a host plant for the overwintering of potato virus M (Perry and McLane 2011), tomato Pepino mosaic virus (Stobbs, Greig, 2014) and potato psyllid Bactericera cockerelli, as a vector for bacteria Candidatus liberibacter solanacearum, which caused the “Zebra chip” disease (Murphy et al., 2013). In addition, several potato and tomato pathogenic fungi were also reported for Solanum dulcamara plants: Boeremia lycopersici (Cooke) Aveskamp, Gruyter et Verkley (= Ascochyta lycopersici Brunaud) in USA (Anonymus, 1960), B. exigua (Desm.) Aveskamp, Gruyter et Verkley (= Phoma exigua var. exigua Desm.) in Poland (Mułenko et al., 2008), Phytophthora infestans (Elansky et al., 2015; Deahl et al. 2010), and Verticillium dahliae Kleb. in Canada (Ginns, 1986).
Spores of many fungal species affecting wild solanaceous plants (e.g., Cladosporium spp. and Alternaria spp.) may provoke allergic reactions in humans. An increased concentration of airborne spores may lead to various disorders of sensibilized persons (Gabriel et al., 2016; Sindt et al., 2016).
The purpose of this study was the investigation of mycobiota associated with live Solanum dulcamara leaves. To improve the identification of fungal species from affected leaf samples, two approaches were used: the isolation of fungi followed by axenic culture analysis and the cloning of species-specific DNA regions in Escherichia coli and further clone library sequencing.
MATERIALS AND METHODS
Solanum dulcamara leaves with visible affection symptoms were collected in 2015 and 2017 (August – September) on the territory of the Lomonosov Moscow State University (Vorobyovy gory). The sampling area (Botanical garden) represented an upland surrounded by multi story houses and located within 7 km from the center of Moscow city. The nearest potato and tomato plantations were located 17 km away from the sampling area, small kitchen gardens – 10 km away.
Twenty leaves with necrotic lesions were sampled from several plants. Sixteen leaves were instantly placed into moist chambers for the isolation of fungi and axenic culture analysis and four leaves were frozen at –75°С for molecular genetic analysis.
Each leaf sample was placed upside down into a moist chamber consisting of a Petri dish with moistened filter paper. After 2–3 days of incubation at 22–25°С, leaves were microscoped. Fungal conidia, sclerotia or mycelia fragments were removed using a fine sterile dissecting needle and transferred to the center of a Petri dish with oatmeal agar supplemented with benzylpenicillin sodium salt (1000 U/ml) to prevent bacterial growth. Cultures were incubated at 22°C until the diameter of a fungal colony reached 25–30 mm. Then a mycelia fragment from the edge of colony was collected with a sterile needle and placed into a fresh Petri dish with an agar medium. The obtained fungal strains were stored in tubes with slant agar at 5°C. Fungal species were identified based on a morphology and micromorphology of cultures or and by sequencing of species-specific genome regions. To analyze axenic fungal cultures by sequencing, ITS5 and ITS4 primers were used according to White et al. (1990). Morphological features of fungal species were compared with the descriptions given in handbooks (Simmons, 2007; Seifert et al., 2011) or with the data from the Q-bank Fungi database (http://www.q-bank.eu/Fungi/).
Leaflets were obtained from the same plants from Botanical Garden. Leaflets were placed on moist chambers, adaxial side up. Leaflets remained unwounded. For sporulating species 3 spore suspensions (10 000 spores/ml) drops of 10µl each were placed per leaflet. For non-sporulating species leaves were infected by mycelium from axenic cultures. Negative controls were drops of sterile distillated water. Petri dishes were kept at room temperature (22°C) for 7 days for disease development (14 h daylight). Lesion development and size were assessed at this point. The experiment was carried out in three replicates.
Total DNA was extracted from each of affected leaves according to the standard CTAB protocol (Kutuzova et al 2017). Amplified rDNA fragments included a part of the 18S gene region, internal transcribed spacer ITS1, 5.8S gene, internal transcribed spacer ITS2, and part of a 28S gene region. Universal ITS1f и ITS4 primers (CTTGGTCATTTAGAGGAAGTAA/TCCTCCGCTTATTGATATGC) were used (White et al., 1990; Gardes, Bruns, 1993). PCR was carried out using commercial PCR core kits (Laboratoriya Izogen Ltd, Moscow, Russia). The following PCR conditions were used: one cycle for 3 min at 96°C, 30 cycles with 30 sec at 94°C, 30 sec at 55°C, 30 sec at 72°C, and 3 min at 72°C. The electrophoretical separation of PCR products was carried out in a TBE buffer and standard 1.5% agarose supplemented with 0.5 µg/ml ethidium bromide as a fluorescent tag. The gels were visualized under a UV light. Amplicons of the required length were extracted from gels using a CleanUp kit (Evrogen Ltd, Moscow, Russia), then inserted into a pAL-TA vector (Evrogen Ltd.) and used for the transformation of Escherichia coli (Dh5a strain) cells, in accordance to Inoue et al. (1990). The resulting clone library was examined by restriction analysis of the amplified DNA insertion using MspI restriction endonuclease (Fig. 1). Based on the obtained results, different restriction insertion profiles were selected. The plasmid DNA was extracted from a sample selected according to the Lee and Rasheed protocol (1990), and nucleotide insertions were sequenced.
Fig. 1.
Electrophoregram of the restriction analysis of clones produced from the sample 4: 1 – Cladosporium herbarum, 2 – Thielavia basicola, 3 – Colletotrichum acutatum, 4 – Phoma herbarum, 5, 10 – Mycosphaerella sp., 6 – Aureobasidium pullulans, 8, 9, 11 – Alternaria alternata, 12 – unidentified Pezizomycotina. M – DNA marker (100–1000 bp).
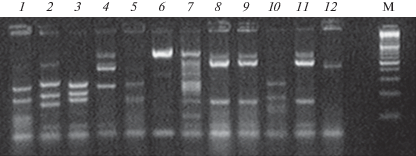
Sequencing was carried out by the Evrogen company (Moscow, Russia). Plasmid DNA was sequenced using a BigDye®Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, CA, USA) and an Applied Biosystems 3730 × l automatic sequencer (Applied Biosystems, CA, USA). The resulted nucleotide sequences were used for the species identification by a comparison with the GenBank databases using BLASTn software.
RESULTS
Oomycete and fungal species isolated from the leaves of bittersweet nightshade by a culture dependent method included Alternaria alternata (Fr.) Keissl, Boeremia exigua, Botrytis cinerea Pers., Coniothyrium fuckelii Sacc., Epicoccum purpurascens Ehrenb., Fusarium oxysporum Schltdl., Phytophthora infestans, Thanatephorus cucumeris (A.B. Frank) Donk.
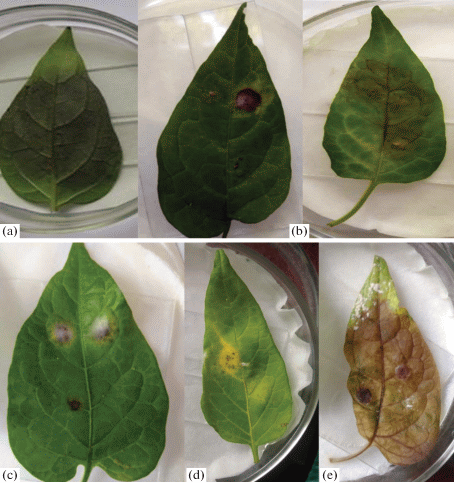
Lesions appeared on leaflets inoculated with Phytophthora infestans, Botrytis cinerea, Alternaria alternata, Epicoccum purpurascens, Rhizoctonia solani (Fig. 2). Of the 3 strains of Boeremia exigua, only one strain caused the appearance of chlorotic spots. First infection symptoms (necrosis) was observed on the third day after inoculation. Coniothyrium fuckelii, Cladosporium sp., Fusarium oxysporum don’t create lesions.
Fig. 2.
Detached leaf assay. Lesions caused by Phytophthora infestans (a), Botrytis cinerea (b), Alternaria alternata (c), Epicoccum purpurascens (d), Thanatephorus cucumeris (e).
The following 14 fungal and oomycete taxa were revealed via the cloning and further sequencing of DNA isolated from bittersweet nightshade leaves: Alternaria alternata (Genbank accession number KU366278), Aureobasidium pullulans (de Bary) G. Arnaud (KU366277), Cladosporium cladosporioides (Fresen.) G.A. de Vries (KU366279), Cladosporium herbarum (Pers.) Link (KU366276), C. tenuissimum Cooke (KU182495), Colletotrichum acutatum J.H. Simmonds (KU366280), C. gloeosporioides (Penz.) Penz. et Sacc., Cryptococcus sp., Mycosphaerella sp., Phialophora sp., Phoma herbarum Westend. (KU366282), Phoma sp., Phytophthora infestans, Thielavia basicola Zopf (KU366283). Alternaria alternata and Phytophthora infestans were revealed by both culture-dependent and DNA-based techniques.
We have failed to identify 7 clones up to the species level. The closest similarity with the NCBI database reference sequences was found for the Pezizomycotina subdivision. However, nucleotide sequences within these clones were different. The phylogenetic analysis allowed us to split them in two distinct groups, one included six clones with similar sequences and another was represented by only one clone.
Each leaf sample analyzed by a cloning procedure hosted a number of fungal and/or oomycete taxa, which often co-occurred within the same infection lesion (Table 1).
Table 1.
Fungal and oomycete species revealed in bittersweet nightshade leaf samples by a cloning procedure
| Leaf sample, No | Number of clones obtained | Number of different restriction profiles | Fungal and oomycete taxa detected |
|---|---|---|---|
| 1 | 11 | 6 | Colletotrichum gloeosporioides, Cladosporium tenuissimum, Cryptococcus sp., Phialophora sp., unidentified Pezizomycotina |
| 2 | 14 | 5 | Alternaria alternata, Cladosporium cladosporoides, Cryptococcus sp., Phoma destructiva, Phytophthora infestans |
| 3 | 18 | 6 | Cladosporium cladosporioides, Cladosporium herbarum, Cryptococcus sp., Phoma sp., Thielavia basicola, unidentified Pezizomycotina |
| 4 | 13 | 8 | Alternaria alternata, Aureobasidium pullulans, Cladosporium herbarum, Colletotrichum acutatum, Mycosphaerella sp., Phoma herbarum, Thielavia basicola, unidentified Pezizomycotina |
DISCUSSION
According to the authors’ knowledge, this was the first complex study of mycobiota inhabiting the leaves of Solanum dulcamara plants growing far from fields of agricultural solanaceous crops. Using cloning (4 leaf samples) and culture-dependent techniques (16 samples), we revealed 18 fungal taxa. Among the species revealed, Boeremia exigua, Alternaria alternata, Botrytis cinerea, Fusarium oxysporum and Thanatephorus cucumeris were known to be typical pathogens of solanaceous plants, which infect potato and tomato plants in Russia. Some of the identified fungal taxa were not typical for these crops or have not been observed in Russia yet. Coniothyrium fuckelii was revealed on potato plants in Poland (Mułenko et al., 2008). Yeast-like Aureobasidium pullulans is known as epiphyte or endophyte of a wide range of plant species (Andrews et al., 2002). It was reported as a tomato pathogen in Brazil and USA (Mendes et al., 1998) and also was isolated from potato leaves with clear blotch symptoms in the Leningrad Region of Russia (Gannibal, 2007).
Colletotrichum acutatum had a worldwide distribution and was pathogenic for a wide range of crops. The species was reported for such host plants as Solanum lycopersicum (Vichova et al., 2012), Capsicum annuum (Jelev et al., 2008), and S. betaceum (Jones, Perez, 2012). Colletotrichum gloeosporioides was also known to be a common plant pathogen found on both leaves (McKenzie, 2013) and fruits (Weir et al., 2012) of Solanum melongena. Phialophora species, which could be either pathogens or saprotrophs (Barnett, Hunter, 1972), were observed on solanaceous crops. For example, P. richardsiae (Nannf.) Conant and P. parasitica Ajello, Georg et C.J.K. Wang were revealed on tomato plants in Poland (Mułenko et al., 2008) and on potato plants in Greece (Thanassoulopoulos, Giapanoglou 1994), respectively. Thielavia basicola was a fungus pathogenic for more than 200 plant species (Shew, Meyer, 1992). It was found on tomato plants in Italy (Venturella, 1991) and has possibly represented the main cause for tobacco stem and root rot in China (Tai, 1979).
A number of plant pathogenic fungi might be harmful not only for plants but also for animal and human health. According to some data, several fungal species revealed during our work (Alternaria alternata, Aureobasidium pullulans, species of the genera Cladosporium, Cryptococcus, Mycosphaerella, and Phoma), might provoke allergic reactions (Adkinson et al., 2013). Alternaria species were able to produce some toxins (altertoxin I, tentoxin, tenuazonic acid) and mutagenic compounds (alternariols) dangerous for humans (Scott et al., 2012).
The co-occurrence of several fungal species in a single affected leaf was noteworthy. Each of the four examined leaf samples harbored 5–8 different fungal species, i.e., complex infections predominated in the affected plant tissues. Some other researchers also reported the same phenomenon. Even low concentration of Alternaria tenuissima inoculum, accompanied by A. solani conidia, promoted the affection of plant tissues (Orina, 2011). There was also data confirming the pathogenicity of A. solani combined with A. alternata (Leiminger, Hausladen, 2011). Associations of A. tenuissima, A. dumosa, A. arborescens, A. infectoria and A. interrupta were observed and examined in Iran (Ardestani et al., 2010). A high pathogenicity of microorganism complexes was also reported for other host plants. Simultaneous co-inoculation of legume plants with leaf-attacking fungal species Mycosphaerella pinodes (Berk. et A. Bloxam) Vestergr. and Phoma medicaginis var. pinodella (L.K. Jones) Boerema impeded the disease progress and reproduction of the fungi (Le May et al., 2009). However, a consecutive inoculation with these pathogens significantly increased the intensity of the disease. Co-occurrence of several fungal species on a single leaf or even within the same necrotic spot did not necessary mean that an “all-at-once” colonization occurred. The infection could be started by a single biotrophic species and then subsequently accompanied by other hemibiotrophic or necrotrophic species.
Thus, 18 different fungal and oomycete taxa were revealed on Solanum dulcamara leaves, and most of them were pathogens of Solanaceae plants. Some of the species revealed have not been earlier reported as the pathogens of solanaceous plants in Russia, but were reported in other countries. In addition, some fungi provoking allergic reactions in immunocompromised humans were identified among the revealed species.
The work was supported by the Russian Foundation for Basic Research under grant N 15-29-02512. The sequencing of all the samples was supported by the Russian Science Foundation under grant N 14-50-00029.
Список литературы
Adkinson N., Bochner B., Burks A. et al. Middleton’s allergy: principles and practice. Philadelphia, Elsevier, 2013.
Andrews J., Spear R., Nordheim E. Population biology of Aureobasidium pullulans on apple leaf surfaces. Can. J. Microbiol. 2002. V. 48 (6). P. 500–513.
Anonymus. Index of plant diseases in the United States. USDA Agricultural Handbook. USA, 1960.
Ardestani S., Sharifnabi B., Zare R., Moghadam A. New Alternaria species associated with potato leaf spot in various potato growing regions of Iran. Iranian J. Plant Pathol. 2010. V. 45. P. 83–86.
Barnett H., Hunter B. Illustrated genera of imperfect fungi. London, Burgess Pub. Co., 1972.
Cooke L.R., Carlisle D.J., Wilson D.G., Deahl K.L. Natural occurence of Phytophthora infestans on woody nightshade (Solanum dulcamara) in Ireland. Plant Pathol. 2002. V. 51. P. 392. https://doi.org/10.1046/j.1365-3059.2002.00705.x
Deahl K.L., Perez F., Baker C.J., Jones R.W. Natural occurrence of Phytophthora infestans causing late blight on woody nightshade (Solanum dulcamara) in New York. Plant Dis. 2010. V. 94. P. 1063. http://dx.doi.org/10.1094/PDIS-94-8-1063B
Elansky S.N., Mita E.D., Kokaeva L.Y. The first record of Phytophthora infestans on Solanum dulcamara in the botanical garden of Moscow State University. Mikologiya i fitopatologiya. 2015. V. 49 (6). P. 386–388 (in Russ.).
Flier W., van den Bosch G., Turkensteen L. Epidemiological importance of Solanum sisymbriifolium, S. nigrum and S. dulcamara as alternative hosts for Phytophthora infestans. Plant Pathol. 2003. V. 52. P. 595–603. https://doi.org/10.1046/j.1365-3059.2003.00922.x
Gabriel M., Postigo I., Tomaz C., Martínez J. Alternaria alternata allergens: Markers of exposure, phylogeny and risk of fungi-induced respiratory allergy. Environment Int. 2016. V. 89–90. P. 71–80. https://doi.org/10.1016/j.envint.2016.01.003
Gannibal Ph.B. Alternariosis of potato leaves – species composition, taxonomy and nomenclature of the disease agents. Vestnik zashchity rasteniy. 2007. V. S. P. 82–93 (in Russ.).
Gardes M., Bruns T. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Molecular Ecol. 1993. V. 2. P. 113–118.
Ginns J. Compendium of plant disease and decay fungi in Canada, 1960–1980. Ottawa, Canadian Government Publishing Centre, 1986.
Inoue H., Nojima H., Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990. V. 6. P. 23–28.
Jelev Z., Bobev S., Minz D., Maymon M., Freeman S. First report of anthracnose fruit rot caused by Colletotrichum acutatum on pepper and tomato in Bulgaria. Plant Dis. 2008. V. 92. P. 172. http://dx.doi.org/10.1094/PDIS-92-1-0172C
Jones R., Perez F. First report of anthracnose caused by Colletotrichum acutatum on tamarillo in the United States. Plant Dis. 2012. V. 96 (4). P. 587. https://doi.org/10.1094/PDIS-09-11-0765
Kutuzova I.A., Kokaeva L.Y., Pobedinskaya M.A., Krutyakov Y.A., Scolotneva E.S., Chudinova E.M., Elansky S.N. Resistance of Helminthosporium solani strains to the fungicides applied for tuber treatment. J. Plant Pathol. 2017. V. 99 (3). P. 635–642. http://dx.doi.org/.10.4454/jpp.v99i3.3950
Le May C., Potage G., Andrivon D., Tivoli B., Outreman Y. Plant disease complex: antagonism and synergism between pathogens of the Ascochyta blight complex on pea. J. Phytopathol. 2009. V. 157. P. 715–721. https://doi.org/10.1111/j.1439-0434.2009.01546.x
Lee S., Rasheed S. A simple procedure for maximum yield of high-quality plasmid DNA. Biotechniques. 1990. V. 9. P. 676–679.
Leiminger J., Hausladen H. Wirkung verschiedener Fungizide auf den Befall der Dürrfleckenkrankheit (Alternaria spp.) sowie auf den Ertrag der Kartoffel. Gesunde Pflanzen. 2011. V. 63. P. 11–18.
McKenzie E. Colletotrichum gloeosporioides. 2013. http://www.padil.gov.au.
Mendes M., da Silva V., Dianese J. Fungosem plants no Brasil. Embrapa-SPI/Embrapa-Cenargen, 1998.
Mułenko W., Majewski T., Ruszkiewicz-Michalska M. A preliminary checklist of micromycetes in Poland. In: Biodiversity of Poland, 9. Krakau, W. Szafer Institute of Botany, Polish Acad. Sci., 2008.
Murphy A., Rondon S., Jensen A. First report of potato psyllids, Bactericera cockerelli, overwintering in the Pacific Northwest. Amer. J. Potato Res. 2013. V. 90. P. 294–296. http://dx.doi.org/10.1007/s12230-012-9281-0
Orina A. Early blight pathogens species composition of Solanaceae crops in Russia. PhD thesis. St. Petersburg, 2011 (in Russ.).
Perry K., McLane H. Potato virus M in bittersweet nightshade (Solanum dulcamara) in New York State. Plant Dis. 2011. V. 95. P. 619. http://dx.doi.org/10.1094/PDIS-10-10-0768
Q-bank Fungi database. http://www.q-bank.eu/Fungi/.
Scott P., Zhao W., Feng S., Lau B. Alternaria toxins alternariol and alternariol monomethyl ether in grain foods in Canada. Mycotoxin Res. 2012. V. 28. P. 261–266. http://dx.doi.org/10.1007/s12550-012-0141-z
Seifert K., Morgan-Jones G., Gams W. et al. The genera of Hyphomycetes. Utrecht, CBS-KNAW Fungal Biodiversity Centre, 2011.
Shew H., Meyer J. Thielaviopsis. In: L. L. Singleton, J. D. Mihail, and C. M. Rush (eds.), Methods for research on soilborn ephytopathogenic fungi. St. Paul, The American Phytopathological Society, 1992. P. 171–174.
Simmons E. Alternaria: An identification manual. Utrecht, CBS-KNAW Fungal Biodiversity Centre, 2007.
Sindt C., Besancenot J., Thibaudon M. Airborne Cladosporium fungal spores and climate change in France. Aerobiologia. 2016. V. 32. P. 53–68. http://dx.doi.org/10.1007/s10453-016-9422-x
Stobbs L., Greig N. First report of bumblebee (Bombus impatiens Cresson) transmission of Pepino mosaic virus between tomato (Solanum lycopersicum L.) and perennial climbing nightshade (Solanum dulcamara L.). Can. J. Plant Pathol. 2014. V. 36. P. 529–533. https://doi.org/10.1080/07060661.2014.954625
Tai F. Sylloge fungorum Sinicorum. Peking, Science Press, Academica Sinica, 1979.
Thanassoulopoulos C., Giapanoglou E. Two new and unusual dry rots of stored potatoes in Greece. Plant Dis. 1994. V. 78. P. 924.
Venturella G. A check-list of Sicilian fungi. Bocconea. 1991. V. 2. P. 5–221.
Víchová J., Stanková B., Pokorný R. First report of Colletotrichum acutatum on tomato and apple fruits in the Czech Republic. Plant Dis. 2012. V. 96. P. 769. https://doi.org/10.1094/PDIS-10-11-0849-PDN
Weir B., Johnston P., Damm U. The Colletotrichum gloeosporioides species complex. Stud. Mycol. 2012. V. 73. P. 115–180. https://doi.org/10.3114/sim0011
White T., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications. 1990. V. 18(1). P. 315–322.
Еланский С.Н., Мыца Е.Д., Кокаева Л.Ю. (Elansky et al.) Обнаружение Phytophthora infestans на Solanum dulcamara на территории ботанического сада МГУ // Микология и фитопатология. 2015. Т. 49. № 6. С. 386–388.
Ганнибал Ф.Б. (Gannibal) Видовой состав, таксономия и номенклатура возбудителей альтернариоза листьев картофеля // Вестник защиты растений. 2007. Т. S. С. 82–93.
Орина А.С. (Orina) Видовой состав возбудителей альтернариоза пасленовых культур на территории России. Автореф. дисс. … канд. Биол. наук. Санкт-Петербург, 2011. 26 с.
Дополнительные материалы отсутствуют.
Инструменты
Микология и фитопатология


