Микология и фитопатология, 2022, T. 56, № 1, стр. 31-35
Xylaria Putaminum (Xylariaceae, Ascomycota), a Rare Mediterranean Species: First Record IN Italy
A. La Rosa 1, V. Agamennone 2, A. Saitta 3, E. Ambrosio 4, *
1 Cooperativa Silene
90100 Palermo, Italy
2 TNO
3704 HE Zeist, The Netherlands
3 Department of Agricultural, Food and Forest Sciences, University of Palermo
90128 Palermo, Italy
4 Via Calamandrei
53035 Monteriggioni Siena, Italy
* E-mail: info@silenecoop.org
Поступила в редакцию 12.05.2021
После доработки 19.09.2021
Принята к публикации 22.10.2021
- EDN: BIQZZY
- DOI: 10.31857/S0026364822010081
Аннотация
Xylaria putaminum is reported for the first time in Italy. The species was described from Northern Africa and this Italian record represents the second one in Europe. The ascomata were collected in the Natural Reserve of “Capo Rama” (Sicily) on senescent fruits of Olea europaea. Macro- and micromorphological descriptions are provided and new information on the geographic distribution and molecular data of this species has now been increased.
INTRODUCTION
The genus Xylaria Hill ex Schrenk (Xylariaceae, Ascomycota) includes 670 saprotrophic inter- and intraspecific taxa (www.mycobank.org) and it is worldwide distributed, especially in tropical and subtropical regions (Kirk et al., 2008; Fournier et al., 2011). Only 16 species of Xylaria have been reported in Italy (Onofri et al., 2005; Saitta et al., 2011; Venturella et al., 2011) and three of those were collected in Sicily (Venturella et al., 2000, 2001). Xylaria putaminum, described by Bertault (1984) for Morocco, has been recently reported for the first time in Europe, in Spain (Sánchez Iglesias, 2016). Therefore, the distributional area, considering the exclusive substrate of growth (on Olea europaea L. fruits), should be considered the Mediterranean basin, as also supported by Fournier (2014) and Sánchez Iglesias (2016).
The substrate of growth for Xylaria species is mainly represented by wood, litter, dead fruits, leaves, seeds, animal dung (Rogers et al., 2005; Hsieh et al., 2010). Many Xylaria species are host-specific, especially when they grow on seeds and fruits (Rogers, 1979; Læssøe, Lodge, 1994; Xu, 1999; San Martín et al., 2001; Rogers et al., 2002). However, the identification can be sometimes problematic since only theleomorphic stromata bearing fertile perithecia can provide enough information for a correct identification (Fournier, 2014).
Since this species has been recorded only in few areas in the world, and, to the best of our knowledge, not before in Italy, the present study aims to increase the knowledge on its geographical distribution and molecular data.
MATERIALS AND METHODS
Study site. The study site is located in the Natural Reserve of “Capo Rama” (38°8′31.43″N, 13°4′10.20″E, Fig. 1) in the locality Terrasini – Province of Palermo, in Sicily (Italy). The sampling site, placed at 34 m a.s.l., was an olive grove dominated by the presence of O. europaea with a relevant accumulation of leaves and fruits. Underneath the olive trees we observed the presence of several ruderal species of plants, being Urtica membranacea Poir., Oxalis pes-caprae L. and Lobularia maritima (L.) Desv the most dominant.
Fig. 1.
Geographic localization of the study site and the world distribution of Xylaria putaminum. Localities of collection are marked with a black circle.
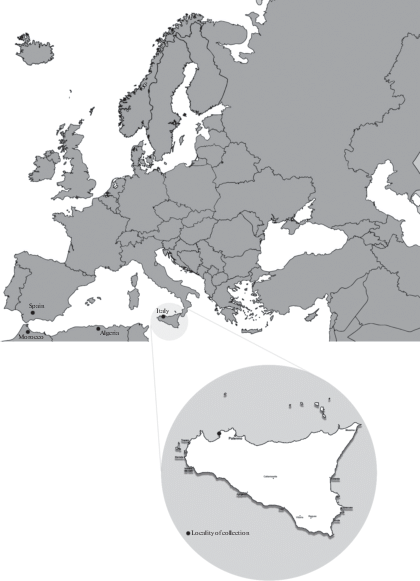
The climate of the area is ascribed to the Mediterranean pluviseasonal oceanic type (Rivas-Martinez 2008). The mean monthly temperature is 25.4°C in August and 11.6°C in January; while, the monthly rainfall varies from 85 mm in December to 2 mm in July (www.climate-data.org).
Survey and Species Identification. The Xylaria putaminum ascomata were collected on senescent fruits of Olea europaea in Autumn 2016 (October–December). The identification and description of the ascomata were based on the macro- and microscopic analysis on fresh and dried specimens. Fresh material was dried in an air dehydrator at 40°C for 30 hours. Microscopic observations and measurements of at least 50 ascospores, 20 asci and 20 paraphyses for each dried specimen, were made using an OPTIKA B-500Tpl microscope and OPTIKAM Pro 5 camera, after rehydrating sections in Congo Red.
The specimens were described following Bertault (1984). Nomenclature and author abbreviations were used in accordance with Bertault (1984) and MycoBank (http://www.mycobank.org).
All voucher specimens are deposited at Herbarium of Natural History Museum, University of Tartu, Estonia (TU).
Molecular analysis. Genomic DNA was extracted in a lysis buffer [0.8M Tri-HCl, 0.2M (NH4)2SO4, 0.2% w/v Tween-20] (Soil BioDyne, Tartu, Estonia) using proteinase K method (100 µl lysis buffer and 2.5 µl proteinase K; incubation at 56°C for 24 h and at 98°C for 15 min). The universal primers ITS5 and ITS4 (White et al. 1990) were used to recover the full ITS that covers roughly 1000 bp. PCR was carried out in 25 µl containing 0.5 µl of each primer, 5 µl FirePol Mastermix (Solis BioDyne, Tartu, Estonia), 1 µl of 10× diluted DNA template and sterilized distilled water. For amplifying the ITS region, PCR conditions consisted of 15 min at 95°C, 35 cycles of 30 sec. at 95°C, 30 sec at 55°C, 1 min at 72°C, and 10 min at 72°C. Amplification success was checked with 1% agarose gel via electrophoresis. PCR products were cleaned using Exo-SAP enzymes GE Healthcare (Freiburg, Germany) using incubation at 37°C for 45 min and at 85°C for 15 min. PCR products were sequenced in Macrogen Inc. (Amsterdam, The Netherlands) using the PCR primers. Quality check and manual editing was performed using Sequencher 5.1 (Gene Codes Corporation, Ann Arbor, USA). The obtained sequence has been deposited to the NCBI GenBank database under the accession number MW750754.
Phylogenetic analysis. The sequence obtained by amplification of the ITS gene was used as a query in BLAST. Highly similar sequences were found through BLASTN searching the nucleotide collection database. Sequences assigned to Xylaria species were extracted and aligned using MUSCLE version 3.8.21_2 (Edgar, 2004). The alignment was curated using trimAl version 1.4.1 (Capella-Gutiérrez et al., 2009) and a tree was inferred with FastTree version 2.1.10_1 (Price et al., 2009). To ensure the validity of the results, both the EMBL-EBI Sequence Analysis Tools (https://www.ebi.ac.uk/services; Madeira et al., 2019) and the NGPhylogeny analysis tools (https://ngphylogeny.fr/; Lemoinel et al., 2019) were used to execute the analytical workflows. The final tree was visualized using the iTol web tool version 5 (https://itol.embl.de/; Letunic, Bork, 2019).
RESULTS
As a result of the macro- and microscopic examination, and molecular analysis, the species Xylaria putaminum was identified. The detailed morphological characteristics of the specimens are provided below.
Taxonomy
Xylaria putaminum Maire et Durieu (Fig. 2).
Fig. 2.
Xylaria putaminum: anamorphic stromata (a); teleomorphic stromata (b); conidiospores (c); ascospores (d).
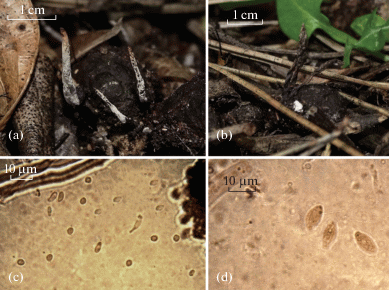
Stromata anamorphic: erected, 0.5–3 ×10–30 mm. Stipes are sterile, fibrillose, blackish with violet hues, 5–20 × 0.5–3 mm, often expanded over the substrate as rhizomorphic. Apical stromata surface is whitish due to numerous conidiophores, which are large, spatuliform and bifurcate. Stipe up to 1/3 or 2/3 of the total stroma length.
Stromata teleomorphic: fusiform, erected, fibrose, 18–39 × 2–5 mm, often unbranched, sometimes with the enlarged, spatuliform or bifurcate apex. Stipes are 5–20 × 0.5–1.4 mm, often sinuous, rarely bifurcate; the surface is longitudinally striate, fibrillose, brown or brown-reddish, sometimes with violet hues, with an enlarged base in correspondence of the substrate of growth. Clavulae are fertile, clearly larger than the stipe, fusiform, narrower at the apex, 8–16 × 0.9–3.5 mm. Apical stromata surface have greyish hues, are brown to black, and are longitudinally striate. Asci are cylindrical and measure (100) 120–150 × (5) 6–7 (9) μm. Paraphyses are abundant, filiform, acuminate, longer than asci, hyaline. Ascospores are ellipsoid, (8) 10–11 (12) × (3) 4–5 μm, smooth, often guttulate, with broadly rounded ends. Conidiospores are amygdaliform, hyaline, and measure (4) 5 (6) × 2 (3) μm.
Material examined: Italy, Sicily, Terrasini, Natural Reserve of Capo Rama, 31.10.2016 (TU124298), 12.11.2016 (TU124299), 01.12.2016 (TU124300), 20.12.2016, 38°8′31.43″N, 13°4′10.20″E – 34 m. a.s.l. On senescent fruits of Olea europaea (TU124830).
Phylogeny
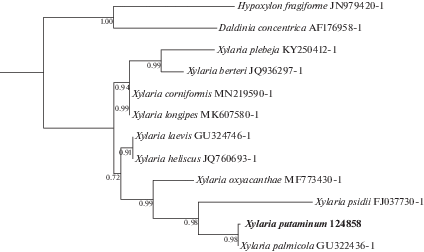
The phylogenetic relationship of Xylaria putaminum with other Xylariales is illustrated in Fig. 3. X. putaminum and X. palmicola form a monophyletic clade with high support value.
Fig. 3.
Phylogenetic relationship of Xylaria putaminum with other Xylaria species, as inferred from ITS nrDNA sequence data. Hypoxylon fragiforme and Daldinia concentrica, two species commonly found on dead and decaying wood, were used as outgroups. Taxon names are followed by the GenBank accession numbers of the sequences. Bootstrap values are indicated on the tree nodes.
DISCUSSION
Xylaria putaminum was collected in Africa (in Algeria), and macromorphological described for the first time in 1920, as reported by Sánchez Iglesias (2016). Later, this species was re-collected (in 1960) in Morocco (Bertault 1984) and it was given a complete morphological description.
Taxonomically, this species belongs to Fusiformes-group because fusoid habitus of its stromata with acute and sterile apex (Bertault, 1984). X. putaminum is well differentiated from other species of Fusiformes by its exclusive substrate of growth: the senescent fruits of Olea europaea. Within the Fusiformes, Xylaria bulbosa (Pers.) Berk. et Broome differs from X. putaminum for the base of the stipe, typically thickened. X. apiculata Cooke has stromata that are brown-argillaceous; X. carpophila (Pers.) Fr. typically grows on decaying fruits of Fagus sylvatica L. while X. oxyacanthae Tul. et C. Tul. grows on fruits of Crataegus spp. The stromal thickness of both X. scopiformis (Mont. ex P. Joly) T. Schumach. and X. pumila (Fr.) Mont. is less than 2 mm, while that of X. juruensis is 2.0–2.5 mm and of X. cupressoides Bertault up to 4 mm (Bertault, 1984; Fournier, 2014). In the context of olive-inhabiting хylarias, it is worth noting the species X. oleagina Thum. described growing on olive fruits and collected in Dalmatia (Thumen, 1883).
Current references on the geographic distribution of X. putaminum point out its presence only in few areas in the word: in Africa (in Marocco and in Algeria, Bertault, 1984) and in Europe (in Spain, Sánchez Iglesias, 2016). Hence, our finding increases the knowledge on the geographic distribution of this species and, to the best of our knowledge, this can be considered the first record in Italy since no other records are available (Onofri et al., 2005; Saitta et al., 2011; Venturella et al., 2011), and the second one in the European continent.
Ecologically, both the African and the Spanish samples were collected on senescent fruits of Olea europaea and O. europaea var. sylvestris, respectively. By our record it possible to confirm the preferential substrate of growth and to extend the distributional area of Xylaria putaminum of more than 1000 Km to the eastern part of the Mediterranean basin (Fig. 1). The distribution of this species is probably underestimated considering that Olea europaea grows in the entire Mediterranean basin (Fournier, 2014).
CONCLUSION
The collection and the identification by both morphological and molecular analysis of Xylaria putaminum allow us to expand our knowledge on the geographic distribution of this species. In addition, the collection of a rare fungal species in Sicily, confirms previous studies on the presence of a high macrofungal diversity in this region (Onofri et al., 2005; Saitta et al., 2011, 2017; Venturella et al., 2011). Future studies should be addressed to investigate less or never studied areas in order to increase our knowledge on the macrofungal species diversity and distribution.
Molecular analysis was performed in the Institute of Ecology and Earth Sciences, University of Tartu, Estonia. The authors would like to thank Leho Tedersoo and the laboratory staff for the support in the molecular analysis.
Список литературы
Bertault R. Xylaires d’Europe et d’Afrique du nord. Bull. Soc. Mycol. France. 1984. V. 100 (2). P. 139–175.
Capella-gutiérrez S., Silla-Martínez J.M., Gabaldón T. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009. V. 25. P. 1972–1973. https://doi.org/10.1093/bioinformatics/btp348
Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004. V. 32. P. 1792–1797. https://doi.org/10.1093/nar/gkh340
Fournier J. Update on European species of Xylaria. 2014. http://www.ascofrance.fr/uploads/xylaria/201406.pdf
Fournier J., Flessa F., Peršoh D. et al. Three new Xylaria species from southwestern Europe. Mycol. Progress. 2011. V. 10. P. 33–52. https://doi.org/10.1007/s11557-010-0671-8
Hsieh H.M., Lin C., Fang M. et al. Phylogenetic status of Xylaria subgenus Pseudoxylaria among taxa of the subfamily Xylarioideae (Xylariaceae) and phylogeny of the taxa involved in the subfamily. Molec. Phylogenet. Evol. 2010. V. 54 (3). P. 957–969. https://doi.org/10.1016/j.ympev.2009.12.015
Læssøe T., Lodge D.J. Three host specific Xylaria species. Mycologia. 1994. V. 86 (3). P. 436–446. https://doi.org/10.1080/00275514.1994.12026431
Lemoinel F., Correia D., Lefort V. et al. NGPhylogeny.fr: new generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019. V. 47. P. 260–265. https://doi.org/10.1093/nar/gkz303
Letunic I., Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019. V. 47. P. 256–259. https://doi.org/10.1093/nar/gkz239
Madeira F., Park Y.M.V., Lee J. et al. The EMBL-EBI search and sequence analysis tools APIs. 2019. V. 47. P. 636–641. https://doi.org/10.1093/nar/gkz268
Onofri S., Bernicchia A.R., Filipello V. et al. Checklist dei funghi italiani. Carlo Delfino Editore, 2005.
Price M.N., Dehal P.S., Arkin A.P. FastTree: computing large minimum evolution trees with profiles instead of a distant matrix. Molec. Biol. Evol. 2009. V. 26. P. 1641–1650. https://doi.org/10.1093/molbev/msp077
Rivas Martinez S. Global Bioclimatics. 2008. http://www.globalbioclimatics.org/book/bioc/global _bioclimatics-2008_00.htm
Rogers J.D. Xylaria magnoliae sp. nov. and comments on several other fruit-inhabiting species. Can. J. Bot. 1979. V. 57. P. 941– 945. https://doi.org/10.1139/b79-115
Rogers J.D., Ju Y.M., Lehmann J. Some Xylaria species on termite nests. Mycologia. 2005. V. 97. P. 914–923. https://www.zobodat.at/pdf/Sydowia_54_0091-0097.pdf
Rogers J.D., San Martín F., Yu Y.M. A reassessment of the Xylaria on Liquidambar fruits and two taxa of Magnolia fruits. Sydowia. 2002. V. 54 (1). P. 91–97.
Saitta A., Bernicchia A., Gorjòn S.P. et al. Biodiversity of wood-decay fungi in Italy. Plant Biosystems. 2011. V. 145 (4). P. 958–968. https://doi.org/10.1080/11263504.2011.633114
Saitta A., Losi C., Ambrosio E. First record of Phlebia margaritae (Polyporales, Basidiomycota) in Italy. Nova Hedwigia. 2017. V. 105 (1–2). P. 37–41.
San Martín F., Lavín P., Rogers J.D. Some species of Xylaria (Hymenoascomycetes, Xylariaceae) associated with oaks in Mexico. Mycotaxon. 2001. V. 79. P. 337–360.
Sánchez Iglesias F. Xylaria putaminum (Ascomycota, Xylariaceae) en Sevilla, suroeste de la Península Iberica. Rev. Catal. Micol. 2016. V. 37. P. 65–73.
Thumen F. Die Pilze des Oelbaumes. Beschreibung sammtlicher bis Ende des Jahres 1883 bekannt gewordener, auf dem Oel- oder Olivenbaume, Olea sativa Lin., Vorkommender Pilze. Bolletino della Societa Adriatica di scienze naturali in Trieste. 1883. V. 8. P. 215–244. https://www.biodiversitylibrary.org/item/46069#page /255/mode/1up
Venturella G., Altobelli E., Bernicchia A. et al. Fungal biodiversity and in situ conservation in Italy. Plant Biosystems. 2011. V. 145 (4). P. 950–957. https://doi.org/10.1080/11263504.2011.633115
White T.J., Bruns T., Lee S. et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics – PCR protocols: a guide to methods and application. 1990. V. 18. P. 315–322. https://msafungi.org/wp-content/uploads/2019/03 /February-2013-Inoculum.pdf
Xu A.S. A new species of Xylaria. Mycosystema. 1999. V. 18. P. 137–140.
Дополнительные материалы отсутствуют.
Инструменты
Микология и фитопатология


