Молекулярная биология, 2021, T. 55, № 6, стр. 927-943
Полногеномные дупликации в эволюции, онтогенезе и патологии: резерв сложности и запаса прочности
О. В. Анацкая a, *, А. Е. Виноградов a
a Институт цитологии Российской академии наук
194064 Санкт-Петербург, Россия
* E-mail: olga.anatskaya@gmail.com
Поступила в редакцию 03.03.2021
После доработки 07.05.2021
Принята к публикации 13.05.2021
Аннотация
Полногеномные дупликации (ПГД), или полиплоидия – это удвоение геномов, которое увеличивает количество генетической информации в клетке. ПГД целых организмов встречаются во всех ветвях эукариот и являются движущей силой видообразования, усложнения и адаптаций. ПГД найдены в соматических клетках всех типов тканей, они могут быть результатом нормальных и измененных онтогенетических программ, регенерации, патологических состояний, старения, малигнизации и метастазирования. Несмотря на универсальность ПГД, их функциональное значение, общие свойства и причины повышенной адаптивности недостаточно ясны. Сопоставление полнотранскриптомных данных и сведений из разных областей молекулярной биологии, геномики и молекулярной медицины показало, что полиплоидия как организмов, так и соматических и раковых клеток ассоциирована с рядом общих признаков, которые позволяют понять, какие именно свойства ПГД приводят к возникновению адаптивного фенотипа. Адаптивность ПГД может быть связана с увеличением сложности регуляции сетей и систем передачи сигналов, устойчивости к стрессу, активацией древних эволюционных программ одноклеточности, путей морфогенеза, выживания и продления жизни. В результате стресса возможен сдвиг баланса между клеточным и организменным уровнями контроля регуляции работы генов в сторону приоритета выживания клетки, что может привести к сердечно-сосудистым заболеваниям и раку. Представленные сведения помогают понять, как полиплоидия создает новые фенотипы и почему она является движущей силой эволюции и важным регулятором биологических процессов в соматических клетках, онтогенезе, патогенезе, регенерации и канцерогенезе.
ВВЕДЕНИЕ
Полногеномные дупликации (ПГД), или полиплоидия, представляют собой удвоение геномов, которое увеличивает количество генетической информации в клетке. Этот процесс является фундаментальным дополнением к копированию ДНК, происходящему во время пролиферации клеток. ПГД играют важную роль в эволюции, нормальной физиологии, регенерации, старении, патологии и канцерогенезе [1, 2]. Если ПГД происходит в половых клетках, то потомство становится полностью полиплоидным, если в соматических, то полиплоидия возникает только в отдельных клетках некоторых тканей. В большинстве случаев полиплоидизация необратима и приводит к долгосрочным последствиям как в эволюции (организменная), так и в онтогенезе (соматическая) [1]. Несмотря на универсальность, биологическое значение ПГД на разных уровнях организации и в разных эволюционных масштабах остается неясным [2]. В то же время, выяснение причин накопления геномов в клетках и роли ПГД в регуляции биологических процессов в филогенезе, онтогенезе и патологии может иметь решающее значение для понимания процессов видообразования, адаптации, дифференцировки, регенерации, патогенеза сердечно-сосудистых заболеваний и опухолевой трансформации. В настоящем обзоре представлены сведения об общих свойствах ПГД на разных уровнях организации, включая организмы, нормальные соматические и опухолевые клетки. Кроме того, рассмотрена важность ПГД для медицины, сельского хозяйства и биотехнологии.
РАСПРОСТРАНЕННОСТЬ ПОЛНОГЕНОМНЫХ ДУПЛИКАЦИЙ
Организмы
Фундаментальный закон эволюции (естественный отбор) следует из увеличения количества организмов в результате размножения, что приводит к конкуренции за ресурсы и естественному отбору. Однако рост сложности не следует из увеличения количества организмов. Он зависит от второго фундаментального процесса, ортогонального первому, а именно – удвоения количества информации в результате ПГД (увеличение информации может происходить из-за удвоения не только целых геномов, но и отдельных генов, но в этом обзоре мы рассматриваем только полногеномные дупликации). Сусуму Оно был первым, кто выдвинул эту концепцию [3]. Его основная идея состояла в том, что в результате дупликации дополнительная копия гена освобождается от стабилизирующего отбора, приобретая тем самым эволюционную пластичность. Он уделил особое внимание ПГД, которые полностью сохраняют регуляторные области и исходный баланс дозы генов, тем самым создавая основу для системных эволюционных изменений. В честь Оно гены, возникающие в результате организменной полиплоидии, назвали “онологами” [4].
ПГД на уровне организмов возникают в тех случаях, когда нередуцированные диплоидные гаметы, появление которых связано с нарушением деления клеток, сливаются с образованием полиплоидов [5]. В филогенетической линии сосудистых растений было два раунда древних ПГД (390 и 300 млн лет назад), как и в филогенетической линии позвоночных (оба около 500 млн лет, близко к кембрийскому взрыву) [6, 7]. Кроме того, более поздние полиплоидные штаммы, виды и надвидовые таксоны широко распространены в природе, сельском хозяйстве и аквакультуре. Они часто встречаются у растений, прокариот, протистов, грибов, беспозвоночных и холоднокровных позвоночных [8, 9]. После ПГД возникает репродуктивный барьер с исходным видом, поэтому полиплоидизация рассматривается в качестве способа мгновенного (instant) видообразования [10]. Количественные оценки распространенности полиплоидных видов могут быть только приблизительными, поскольку не у всех существующих видов проведен скрининг на наличие недавних ПГД. Предполагается, что около 35% видов высших (цветковых) растений являются недавними полиплоидами [10]. Интересно, что процент полиплоидных видов увеличивается при движении от экватора к более высоким широтам (что указывает на повышенную стрессоустойчивость полиплоидов). Так, наиболее высокий процент полиплоидов наблюдается в тундре (51%), в тайге их немного меньше (47%), в умеренной зоне еще меньше (38–40%), а в тропиках их менее 20% [10].
У животных можно найти оценки только для отдельных групп. Интересны группы с наибольшим процентом полиплоидов, поскольку они могут рассматриваться в качестве возможного верхнего предела. У рыб такой группой может быть подсемейство карповых (Cyprininae), представленное 1300 видами, из которых около 400 полиплоидные, что составляет примерно 30% (возможно, однако, что не все остальные виды проверены) [11]. У амфибий рекордсменами являются лягушки рода Xenopus, у которых полиплоидны 26 из 27 видов (96%), включая тетраплоидов, октоплоидов и додекаплоидов [12]. Ситуация резко меняется при переходе к амниотам (и далее к теплокровным). Так, у рептилий полиплоиды возникают очень редко, обычно это триплоиды, которые не способны к половому размножению и воспроизводятся только партеногенетически. Таких видов около 0.6% [13]. У птиц и млекопитающих полиплоидные виды вообще не обнаружены. У двух видов птиц описано спонтанное появление жизнеспособных триплоидов, но из-за своей стерильности они не могут образовывать новые виды [14]. Сообщалось о тетраплоидности одного из видов грызунов, однако позднее эти данные были опровергнуты [15].
Причины отсутствия полиплоидии у теплокровных животных пока не ясны. Известно, что полиплоидные эмбрионы млекопитающих гибнут еще на ранних стадиях (в основном на стадии бластоцисты) [16]. Наиболее правдоподобным нам кажется предположение, что причиной их гибели может быть взаимосвязь между полиплоидией и повышенной генетической нестабильностью, которая усиливается при переходе к теплокровности [17–19]. Известно, что теплокровность связана с активацией кислородного обмена и с усилением окислительного стресса, а также с АТР-зависимой декомпактизацией хроматина [20, 21]. Эти свойства могут ускорять накопление повреждений ДНК и лишать клетку способности к нормальному делению [18].
Примечательно, что некоторые модельные организмы являются относительно недавними полиплоидами (дрожжи, данио, ксенопус, арабидопсис) [1]. Полиплоиды могут возникать также в результате аллополиплоидизации, или слияния геномов разных видов [22, 23]. Важно отметить, что закрепление ПГД в эволюции происходит на фоне ряда негативных последствий. Это связано с нарушением коньюгации хромосом в мейозе, повышением вероятности анеуплоидии, эпигенетической нестабильности, избыточности регуляторного аппарата и изменения архитектуры клетки [24]. При этом следы древних геномных дупликаций сохранились практически у всех современных эукариот, что свидетельствует об адаптивных свойствах.
Нормальные соматические клетки
ПГД обнаружены в соматических клетках тканей всех многоклеточных, включая водоросли, мхи, лишайники, сосудистые растения, беспозвоночные и позвоночные животные, что подтверждает эволюционную успешность этого явления [1, 25].
Дупликация геномов, как правило, возникает в результате онтогенетических программ, инициирующих переключение с полного клеточного цикла на цикл незавершенный [26]. В результате происходит разобщение между репликацией ДНК и делением клетки. В зависимости от степени рестрикции последних фаз клеточного цикла это может приводить к эндомитозу или эндоредупликации с формированием полиплоидных клеток [27]. Полиплоидия может быть вызвана также слиянием клеток, например, при образовании синцития в миобластах и трофобласте [28].
У человека и теплокровных животных полиплоидные клетки формируются преимущественно в ходе нормального органогенеза сердца, коры головного мозга (пирамидные нейроны), ретины, печени, поджелудочной железы, плаценты, сосудов, кожи, крови и других органов [29]. Все этих органы и ткани характеризуются низкой способностью к самообновлению и постоянно высокой специфической нагрузкой, которая необходима для непрерывного обеспечения функций.
Как и в случае ПГД, соматическая полиплоидия сохраняется в эволюции на уровне организмов, несмотря на ряд побочных эффектов, включая нестабильность генома, изменение архитектуры клетки и энергетическую затратность поддержания дополнительных геномов, указывая на то, что преимущества полиплоидии перевешивают ее недостатки [30, 31].
Опухолевые клетки
Если ПГД возникают в соматических клетках, которые не запрограммированы на полиплоидизацию и обладают высоким пролиферативным потенциалом, то накопление геномов может способствовать онкотрансформации [32]. Недавнее исследование метастатических солидных опухолей, проведенное на 2520 парах транскриптомов опухолевых и нормальных тканей, выявило полиплоидные клетки в 56% всех образцов [33]. При этом процент полиплоидных клеток варьировал от 15% в опухолях нервной системы до 85% в опухолях пищевода [33]. Кроме того, обнаружена положительная корреляция между содержанием полиплоидных клеток в опухолях молочной, поджелудочной и щитовидной железы, глиобластомах, колоректальной карциноме, а также в опухолях костной ткани, почек, яичников, предстательной железы и другой локализации со степенью злокачественности, устойчивостью к терапии, рецидивами и плохим прогнозом [34, 35].
Опухолевые полиплоидные клетки формируются в результате запуска программ выживания в трудных условиях и адаптации к стрессу [36]. Эти программы могут проявляться в виде выпадения последних фаз клеточного цикла, преждевременного вхождения в клеточный цикл, нерасхождения хромосом и многополюсности митоза [37–39]. Гигантские полиплоидные опухолевые клетки могут появляться путем слияния клеток, пульверизации хромосом (хромотрипсиса) и асинхронии митотических процессов в двуядерных клетках [26]. Удивительно, что полиплоидизация путем слияния различных типов клеток (эпигенетическая гибридизация), которая часто наблюдается при раке [28, 40], напоминает аллополиплоидизацию (генетическую гибридизацию) в эволюции и у сельскохозяйственных культур. Подобно аллополиплоидным организмам, гибридные полиплоидные клетки приобретают стрессоустойчивость (например, химио- и радиационную устойчивость) и адаптивную пластичность [1].
Важно отметить, что общим для нормальной и опухолевой полиплоидии является ряд сопровождающих их изменений, включая аномально большой размер клеток, склонность к митотическим катастрофам, анеуплоидии, изменению метаболизма и повреждению ДНК, а также изменение регуляторных программ, что предполагает существование у полиплоидных клеток адаптивных преимуществ, которые перевешивают их недостатки [37, 41, 42].
ОРГАНИЗМЕННОЙ И КЛЕТОЧНОЙ ПОЛНОГЕНОМНОЙ ДУПЛИКАЦИИ СВОЙСТВЕННА РЕДИПЛОИДИЗАЦИЯ
Редиплоидизация – это превращение полиплоидных наборов хромосом в диплоидные. Редиплоидизация означает не возврат к первоначальному диплоидному состоянию, а лишь восстановление правильной коньюгации хромосом вследствие их перестройки и дивергенции ДНК [43, 44].
Организмы
На уровне организмов редиплоидизация сопровождается потерей генов, субфункционализацией и неофункционализацией [45, 46]. Потеря генов происходит в результате делеции или псевдогенизации (мутационного разрушения гена, который больше не поддерживается естественным отбором). Субфункционализация – это разделение функции исходного гена, когда каждый из дупликатов выполняет только часть исходной функции (или всю функцию, но только в части тканей или стадий развития), тогда как неофункционализация – это приобретение новой функции [47, 48]. Субфункционализация и неофункционализация приводят к дивергенции генов, что должно сопровождаться интеграцией работы растущего числа диверсифицирующих генов [49]. В полногеномных исследованиях субфункционализация может быть выявлена по изменениям экспрессии генов и свойств интерактома (сети белковых взаимодействий). Изменения экспрессии можно обнаружить в тех случаях, когда каждая копия дуплицированного гена начинает экспрессироваться только в части тканей или стадий развития, в которых экспрессировался исходный ген. Изменения интерактома обнаружены у дрожжей. В этом случае белки, кодируемые онологами, имеют меньшее количество взаимодействий по сравнению с недуплицированными генами (и даже по сравнению с генами, появившимися в результате отдельных генных дупликаций, а не ПГД) [50]. Это свидетельствует о том, что субфункционализация была преобладающей тенденцией в эволюции сохранившихся онологов. Функциональный анализ перекрывающихся и неперекрывающихся интерактантов в каждой паре онологов показал, что регуляторные функции гораздо чаще встречаются в неперекрывающихся интерактантах, свидетельствуя о том, что именно регуляторные изменения были важны для дивергенции онологов.
Соматические клетки
В некоторых случаях запрограммированная в онтогенезе полиплоидизация может сменяться запрограммированным распадом клеток. В результате гигантские клетки с множеством геномов могут распадаться на клетки меньшей плоидности c помощью амитоза или редукционного деления [38, 39]. Например, полиплоидные энтероциты дрозофилы при голодании проходят через амитоз и образуют тканеспецифические стволовые клетки [51]. Подобным же образом культивируемые полиплоидные клетки костного мозга и гигантские клетки трофобласта могут разделяться на одноядерные клетки меньшей плоидности [52, 53].
Раковые клетки
Гигантские полиплоидные опухолевые клетки со стволовыми свойствами, как и полиплоидные клетки соматических тканей, способны распадаться на диплоидные потомки [35, 36, 42, 54]. Это явление, как правило, наблюдается в стареющих полиплоидных клетках, выживших после химио- или радиотерапии [55]. Диплоидизация происходит благодаря активации герминативных программ мейоза через включение консервативных регуляторов мейотической профазы оогенеза, например, киназы MOS и генов SPO11, DMC1, RAD51, REC8, OCT4A, VASA и FRAGILIS [35]. После диплоидизации снова наступает полиплоидизация через канонический клеточный цикл с потерей последних фаз деления [35, 36, 42, 54]. Чередование полиплоидных и диплоидных фаз жизненного цикла наделяет опухолевые клетки способностью к бессмертию и сопровождается активацией древних программ одноклеточности и выживания, неотъемлемой части канцерогенеза [35, 56]. Важно отметить, что распад гигантских полиплоидных опухолевых клеток происходит подобно тому, как это бывает у простейших, например, у кишечной амебы [55, 57, 58]. Таким образом, это явление можно рассматривать как аналогию эволюционной редиплоидизации, напоминающей чередование жизненных циклов простейших. При этом важно учитывать, что этот процесс не является возвратом к исходному диплоидному состоянию.
ПОЛНОГЕНОМНЫЕ ДУПЛИКАЦИИ СПОСОБСТВУЮТ УВЕЛИЧЕНИЮ СЛОЖНОСТИ, ВИДООБРАЗОВАНИЮ И ПОЯВЛЕНИЮ КЛЕТОК С НОВЫМИ СВОЙСТВАМИ
Организмы
Увеличение сложности регуляции и систем передачи сигналов – отличительная черта эволюции организмов после ПГД [1, 59]. Регуляторные гены сохраняются после ПГД, поскольку ослабленный стабилизирующий отбор позволяет переформатировать дуплицированные клеточные сети и развить новые функциональные возможности, увеличивающие биологическую сложность [2]. Примечательно, что в интерактоме человека одиночные древние гены (не онологи) имеют больше взаимодействий, чем новые гены, что указывает на постепенный эволюционный рост глобальной сети межгенных взаимодействий от центральной части к периферии [60]. Однако во время основного этапа развития многоклеточной организации от Bilateria до Euteleostomi (костных позвоночных) наблюдается плато, когда число взаимодействий новых генов остается на том же уровне. Это плато связано с серией полногеномных дупликаций и указывает на то, что ПГД удваивают и центральную часть интерактома, которая наиболее богата взаимодействиями. Таким образом, сложность интерактома повышается скорее в результате ПГД, чем дупликации отдельных генов. В дополнение к увеличению сложности полиплоидные организмы обладают повышенной эволюционной пластичностью [61]. Часто за событиями ПГД следует крупномасштабная радиация видов в пределах таксономической группы, что указывает на важность геномных дупликаций для видообразования [62].
Соматические клетки
В соматических клетках ПГД также способствуют возникновению новых признаков. Например, в гепатоцитах, кардиомиоцитах, интерстициальных клетках сердца и стволовых клетках жировой ткани наблюдаются тотальные изменения транскриптома, которые проявляются в основных метаболических и сигнальных путях, включая активацию древних генов и ответа на стресс, изменяются также пути пролиферации, метаболизма и морфогенеза [63–67]. ПГД способствуют также образованию клеток с новой специализацией [68]. В частности, показано, что полиплоидные клетки костного мозга могут быть источником одноядерных диплоидных мезенхимных стволовых клеток, а дифференцированные энтероциты дрозофилы способны превращаться в стволовые клетки кишечника [51, 53]. Интересно отметить эволюционную консервативность феномена распада полиплоидных клеток с образованием диплоидных потомков с новыми свойствами.
Опухолевые клетки
Подобно соматическим клеткам, полиплоидные клетки соматических опухолей приобретают новые характеристики, такие как эмбриональность, способность к миграции, устойчивость к стрессу, перестройка путей пролиферации, метаболизма и ответа на повреждение ДНК [35, 36, 57, 69, 70]. Полнотранскриптомное изучение особенностей контроля клеточного цикла в гигантских полиплоидных клетках рака молочной железы показало, что полиплоидия ассоциирована, с одной стороны, с повышенной экспрессией генов контрольных точек G2/M (по-видимому, это ответ на повреждение ДНК), с другой – с пониженной экспрессией генов контрольных точек G1/S и генов цитокинеза [66]. Это явление характерно и для стволовых клеток [71–73]. Возможно, что избыточная активация контрольной точки G1/S (которая может быть связана с повреждением ДНК) задерживает переход G2/M. Предполагается также, что полиплоидизация опухолевых клеток обусловлена адаптацией к репликативному и метаболическому стрессу [66, 74–76].
Существует еще одна важная особенность полиплоидных опухолевых клеток – проявление древних эволюционных программ и признаков одноклеточности [41, 57, 58]. Атавистическая теория онкогенеза предполагает, что рак является возвратом от многоклеточного состояния к одноклеточному и часто связан с полиплоидизацией [58, 77, 78]. Недавно показали, что при раке повышается экспрессия генных модулей одноклеточного эволюционного происхождения, тогда как экспрессия модулей многоклеточного происхождения снижается [67, 79, 80]. Возможно, это и приводит к распаду полиплоидных гигантов на мелкие клетки со стволовыми свойствами.
Примечательно, что околодиплоидные потомки полиплоидных опухолевых клеток со стволовыми свойствами также проявляют черты эмбриональности, экспрессируют основные маркеры плюрипотентности и способны распространяться далеко за пределы опухоли [35, 36, 81, 82]. В дальнейшем такие клетки могут дифференцироваться в разные типы клеток, которые экспрессируют маркеры эндотелия, фибробластов, макрофагов, эритроцитов, адипоцитов и даже миоэпителиальных клеток [83, 84]. Эти свойства полиплоидных опухолевых клеток согласуются с гипотезой о возможной роли опухоли в эволюции организмов, согласно которой опухоль представляет собой источник дополнительных клеточных масс, экспрессирующих эволюционно новые гены, которые определяют морфологические новшества и эволюционные инновации [85, 86]. Эти свойства показывают также, что полиплоидия в соматических и опухолевых клетках может быть источником клеток нового типа, подобно тому как ПГД организмов способствуют видообразованию.
ПОЛНОГЕНОМНЫЕ ДУПЛИКАЦИИ ПОВЫШАЮТ УСТОЙЧИВОСТЬ К СТРЕССУ
Организмы
Полиплоидам свойственна более высокая устойчивость к болезням и стрессу, чем диплоидам [87, 88]. Полиплоиды особенно адаптивны при резких изменениях среды [1, 89]. Многие эволюционные линии, претерпевшие ПГД, выжили во время глобальных вымираний в отличие от их неполиплоидных предков, что указывает на повышенную адаптивность к стрессовым условиям и экологическим катастрофам [62]. Исследования, проведенные на дрожжах, свидетельствуют о том, что полиплоидия может ускорить эволюционную адаптацию к сложной окружающей среде благодаря пластичности и адаптивности регуляторных сетей [42, 90]. На модельных (дигитальных) организмах показано, что ПГД способствует адаптации к нестабильным и стрессовым условиям, в то время как в благоприятных условиях поддержание ПГД, напротив, снижает адаптивность [89, 91]. Природа преимуществ геномных дупликаций в стрессовых условиях и их недостатков в обычных условиях пока не ясна. Одна из гипотез предполагает, что в стабильной среде случайные мутации в генах регуляторных сетей часто бывают вредными, а в стрессовой среде, напротив, эти изменения повышают пластичность и открывают дорогу радикальным адаптациям, необходимым для выживания [91].
Соматические клетки
В соматических клетках ПГД (как и на уровне организмов) способствует адаптации к стрессу. Изучение кардиомиоцитов разных видов теплокровных показало, что самое большое число геномов характерно для видов, способных выживать в экстремальных условиях [92, 93]. Например, у птиц максимальная плоидность клеток наблюдается у дальних мигрантов – представителей отряда Anseriformes (гусей и уток) [94, 95]. Эти птицы не способны переносить высокие аэробные нагрузки такие, например, как быстрый и маневренный полет, зато они приспособлены к нагрузке, связанной с длительным полетом в условиях гипоксии. При миграции птицы совершают длительные перелеты (по 7–8 ч) на высоте 8–10 км при содержании кислорода около 30% от нормального уровня и при температуре примерно –40°С [94, 95]. Среди млекопитающих самая высокая плоидность кардиомиоцитов характерна для человека, у которого каждая нормальная клетка содержит 4–16 геномов [96]. У человека повышенная нагрузка на сердце связана с прямохождением [96]. Важно отметить, что при гипоксических состояниях, вызванных, например, тетрадой Фалло, гипертонией и ишемией, кардиомиоциты человека могут накапливать до 64 и даже 128 геномов [96–98].
Существование взаимосвязи между ПГД в кардиомиоцитах и стрессом подтверждают и экспериментальные данные. На модели неонатальных крыс показано, что воспалительный стресс, вызванный инфекционным гастроэнтеритом, приводит к избыточной и необратимой полиплоидизации в мышечных клетках сердца и печени [99–103]. Показано также, что преждевременное попадание недоношенных ягнят в кислородную среду приводит к необратимому накоплению одноядерных полиплоидных клеток в правом и левом желудочках сердца [104]. Авторы этих независимых исследований пришли к общему выводу, что стресс во время дифференцировки может привести к избыточному накоплению геномов и к онтогенетическому программированию сердечно-сосудистых заболеваний [99, 104, 105].
Множество других примеров взаимосвязи между соматической полиплоидией и устойчивостью к гипоксическому, осмотическому, окислительному, токсическому, стрессу в разных типах клеток млекопитающих, растений, насекомых и других многоклеточных и одноклеточных эукариот приведены в обзорах [2, 106].
Важно отметить, что ПГД в соматических клетках, как и в организмах, повышают адаптацию к стрессу, но снижают адаптацию к нормальным условиям. Исследования ПГД в гепатоцитах и кардиомиоцитах нескольких десятков видов млекопитающих и птиц с разным функциональным потенциалом сердца и печени показали, что максимальная полиплоидизация гепатоцитов и кардиомиоцитов характерна для животных с небольшим метаболическим диапазоном и низким индексом сердца, а значит, ассоциирована со снижением функционального потенциала органов [95, 107–109]. Показано также, что увеличение количества ДНК в ядре повышает устойчивость конденсированного состояния хроматина к колебаниям ионного состава среды, окружающей клетку [110].
Опухолевые клетки
В клетках опухолей, которые подвергаются стрессу, например, из-за потери кровоснабжения, накопления токсичных метаболитов, изменения pH, иммунной инфильтрации и терапии, ПГД является одним из механизмов адаптации к стрессу [2, 70, 111]. Первые экспериментальные данные, подтверждающие роль стресса в формировании гигантских полиплоидных опухолевых клеток, получены в опытах, проведенных на опухолевых клетках, культивируемых в условиях гипоксии или с применением химических индукторов гипоксии [84, 112]. Позже показали, что полиплоидные опухолевые клетки могут возникать и после воздействия химио- и радиотерапии, механического стресса и вирусной инфекции [68, 113, 114].
Многочисленные исследования свидетельствуют о том, что гигантские полиплоидные клетки особенно характерны для метастатических форм опухолей, резистентных к химио- и радиотерапии [57, 113–115]. Дополнительные геномы усиливают приспособляемость к быстро меняющимся условиям среды благодаря тому, что геномные копии защищают клетку от повреждения ДНК и вредных мутаций и делают адаптационную перестройку регуляторных путей более обширной [111, 115]. Повышенной адаптивности полиплоидных опухолевых клеток способствует также усиленная способность к метастазированию и подвижности, что в некоторых случаях позволяет быстро выйти из неблагоприятных условий, например, из гипоксии [112]. Кроме того, гигантские полиплоиды способны впадать в состояние метаболического покоя (dormancy), что позволяет пережить периоды сильного стресса, связанного, например, с химио- и радиотерапией [113]. После нормализации условий, полиплоидные гиганты распадаются на множество мелких диплоидных или парадиплоидных потомков, которые также обладают высокой адаптивностью [36, 42, 82].
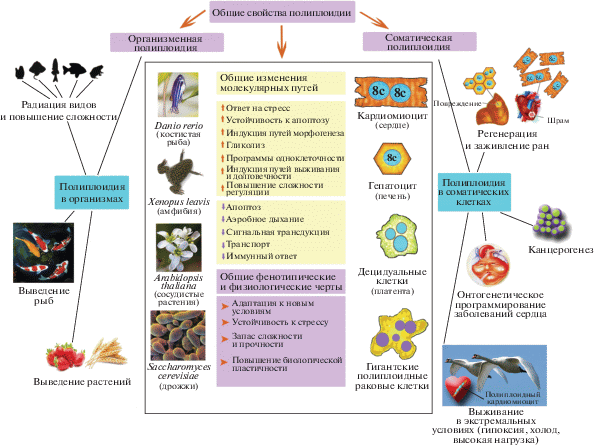
ОБЩИЕ ПРОЯВЛЕНИЯ ПОЛНОГЕНОМНЫХ ДУПЛИКАЦИЙ В БИОЛОГИЧЕСКИХ ПРОЦЕССАХ
Недавние исследования показали, что ПГД могут существенно влиять на транскриптом клеток, генные модули и структуру сетей межбелковых взаимодействий [63, 65, 66, 101, 116, 117]. К наиболее важным проявлениям ПГД относятся активация путей морфогенеза, устойчивость к стрессу и включение древних программ одноклеточности [58, 66, 117], а их иллюстрацией служат биоинформатические данные, полученные с помощью попарно-перекрестного сравнения транскриптомов сердца и печени человека и мыши [101, 116, 117]. Эти пары органов были выбраны потому, что для них характерны сильные различия в полиплоидизации. Так, кардиомиоциты человека имеют полиплоидные ядра, а гепатоциты содержат диплоидные ядра, тогда как у мыши, напротив, кардиомиоциты имеют диплоидные ядра, а гепатоциты – полиплоидные [116]. Средняя плоидность ядер гепатоцитов человека и мыши составляет 2.05 ± 0.01 и 5.47 ± 0.10 геномов, соответственно, в то время как средняя плоидность ядер кардиомиоцитов человека и мыши равна 4.04 ± 0.05 и 2.05 ± 0.01 геномов [30].
Метод попарно-перекрестного сравнения транскриптомов позволил исключить видо- и тканеспецифические эффекты и идентифицировать эффекты полиплоидии. Показано, что полиплоидия ассоциирована с проявлениями морфогенетических программ (пути Notch, TGFb, Hippo, WNT и др.), усилением ответа на гипоксию, активацией гликолиза и снижением активности путей апоптоза, иммунного ответа и аэробного метаболизма [30, 101, 116, 117]. Кроме того, выявлена тесная ассоциация между полиплоидией и усилением протеасомной деградации белка [66, 117, 118], которая также может быть ассоциирована с проявлениями стволовости [119]. Взаимосвязь между полиплоидией и эмбриональностью подтверждают также данные о том, что ПГД способствуют активации интерактома онкогена с-Myc и усилению экспрессии генов из древних филострат, относящихся к одноклеточной стадии эволюции [66, 117]. К проявлениям адаптации к стрессу можно отнести усиленный ответ на повреждение ДНК, а также окислительный, биотический и абиотический стрессы [116].
Результаты исследований, проведенных на мышечных и интерстициальных клетках сердца человека и мыши, энтероцитах дрозофилы и ретиноцитах человека в культуре, подтверждают, что полиплоидия может быть ассоциирована с проявлениями эмбриональности, усилением синтеза ДНК и ответа на стресс [64, 96]. Кроме того, накопилось большое количество сведений о том, что полиплоидия способствует адаптации к окислительному, воспалительному, гипоксическому и генотоксическому стрессу [27, 120].
Причиной изменения активности транскриптома при ПГД могут быть эпигенетические модификации хроматина, связанные с метилированием ДНК, модификациями гистонов и негистоновых регуляторов хроматина, например, HNMGB1 и HMGB2, а также с изменением экспрессии микроРНК [121–125]. Исследования, проведенные на гепатоцитах и кардиомиоцитах, выявили взаимосвязь между полиплоидией и декомпактизацией хроматина [121, 126, 127]. Показано также, что при полиплоидии может возникать изменение баланса антагонистической активности метилтрансферазных комплексов гистонов Trithorax и Polycomb, которые регулируют метилирование гистонов в CpG-богатых областях и модулируют экспрессию многих генных модулей, как это происходит, например, в кардиомиоцитах, децидуальных клетках, гепатоцитах, клетках карциномы в культуре и клетках растений [104, 121, 128, 129].
Недавно обнаружили взаимосвязь между ПГД и повышенной экспрессией бивалентных генов, содержащих активаторные (H3K4me3) и репрессорные (H3K27me3) домены хроматина [66]. Бивалентные гены часто совпадают с генами транскрипционных факторов, вовлеченных в изменение клеточной судьбы, эмбриогенез и канцерогенез [130]. Наделяя клетку способностью к быстрому изменению паттерна экспрессии генов, бивалентные гены также способствуют возникновению адаптационных самоорганизующихся регуляторных сетей [42]. Адаптация генных сетей включает поиск оптимальных состояний клетки, что дает возможность совмещать несовместимые (в диплоидном состоянии) процессы, например, старение и стволовость, аэробное дыхание и анаэробный гликолиз, а также эволюционно древние программы одноклеточности и относительно молодые программы многоклеточности [42]. В результате, полиплоидные клетки обладают более высокой адаптивностью и жизнеспособностью, чем диплоидные [82, 111].
Важно отметить, что основные особенности транскриптома соматических полиплоидных клеток совпадают с транскриптомом фибробластов долгоживущих видов, например, голого землекопа и слепыша, а также у мутантов мыши, нематоды, дрозофилы и дрожжей с повышенной продолжительностью жизни [131–133]. Это, например, ответ на гипоксию, индукция путей репарации, пролиферации, морфогенеза, гликолиза, протеасомной деградации белка, адаптация к стрессу, а также негативная регуляция апоптоза и замедление аэробного метаболизма [27, 134, 135]. ПГД на уровне отдельных клеток увеличивает продолжительность жизни клеток в тканях и в культуре, в основном, из-за их повышенной устойчивости к апоптозу, мутациям и генетической нестабильности [1].
Фундаментальность перечисленных свойств соматической полиплоидии подтверждает то, что ряд аналогичных генных модулей сохранился в дуплицированном состоянии у древних полиплоидов, таких как дрожжи, арабидопсис, аксолотль, ксенопус и костистые рыбы [1, 106]. К таким дуплицированным модулям относятся биогенез рибосом, транскрипция, пролиферация, гликолиз, адаптация к гипоксическому и окислительному стрессу и негативная регуляция апоптоза [106, 136]. Аналогично, генные модули, ингибированные при соматической полиплоидии (аэробное дыхание, сигнальная трансдукция, транспорт, апоптоз и иммунный ответ), у тех же древних полиплоидов содержат гены, утратившие дупликаты. Этот факт предполагает общие свойства между организменной и соматической полиплоидией. Вполне вероятно, что адаптация к стрессу, которая, по-видимому, способствует возникновению соматической полиплоидии, может играть важную роль в эволюционной фиксации полиплоидии на уровне организма.
Таким образом, свойства транскриптома, связанные с полиплоидией, консервативны в филогенезе и онтогенезе и направлены на усиление выживаемости в новых условиях, адаптивности к стрессу и увеличение продолжительности жизни (рис. 1). Вовлеченность геномных дупликацией в регуляцию программ развития, продолжительности жизни и адаптации к стрессу делают особенно важным изучение роли полиплоидии в патологических и физиологических процессах, которые могут затрагивать постнатальный морфогенез и адаптацию, включая онтогенетическое программирование широко распространенных заболеваний, регенерацию тканей и процессы опухолевой трансформации. В связи с этим мы подробнее рассмотрим данные, которые могут прояснить роль полиплоидии в медицине.
КЛИНИЧЕСКОЕ ЗНАЧЕНИЕ ПОЛНОГЕНОМНЫХ ДУПЛИКАЦИЙ
Онологи и болезни
Сохранившиеся онологи человека – это чувствительные к дозе гены [137–139]. Чувствительность к дозе генов вызывает все больший интерес, поскольку может дать ключ к патогенезу многих заболеваний [138]. Модель баланса дозы генов предполагает, что среди всех комплексных генных продуктов в биохимическом пути поддерживается стехиометрическое равновесие, поэтому изменение экспрессии, мутация или вариация числа копий даже в одном гене пути могут быть вредными. Изменения в паттерне экспрессии онологов обнаруживаются при различных типах рака (молочной железы, предстательной железы, толстой кишки, щитовидной железы, яичника, мочевого пузыря, шейки матки, легких, матки) и в ангиогенезе опухолей [139]. Гены моногенных заболеваний человека, представленные в базе данных OMIM, обогащены онологами [138].
Полногеномные дупликации в опухолевых клетках
До недавнего времени полиплоидные опухолевые клетки считали безопасными, потому что их рассматривали как необратимо стареющие клетки, утратившие способность к делению [111]. Лишь относительно недавно показали, что эти многоядерные гиганты обладают стволовыми свойствами и являются важными факторами онкогенности, метастазирования и устойчивости к ядам [58, 140–144]. Полиплоидные клетки присутствуют у пациентов с метастатическими опухолями всех типов, они найдены в опухолевых линиях всех типов [111, 115]. Многочисленные клинические и экспериментальные данные, полученные на разных моделях, подтверждают гипотезу о том, что устойчивость опухолевых клеток к терапии обусловлена появлением в их популяции полиплоидов, и такие клетки могут выживать после химио- и радиотерапии [113, 115, 140]. Многие исследователи считают, что гигантские полиплоидные опухолевые клетки, обладающие повышенной биологической и эпигенетической пластичностью и способностью к метастазированию, являются главными виновниками летальности [115, 145]. К опасным свойствам относят также склонность этих клеток впадать в состояние покоя в ответ на терапию и редиплоидизироваться после нормализации условий микроокружения. Таким образом, полиплоидные опухолевые клетки имеют важное клиническое значение, а разработка подходов к их элиминации представляет важную задачу терапии опухолей.
Полиплоидия при регенерации
ПГД считается важным фактором сохранения функции и выживания клеток при повреждениях [29, 62]. Полиплоидные клетки участвуют в регенерации органов и тканей, состоящих из терминально дифференцированных клеток, где отсутствуют резидентные стволовые клетки, поэтому восстановление с помощью пролиферации диплоидных клеток невозможно [120]. Например, так происходит в сердце человека и млекопитающих, где полиплоидия может быть единственным способом сохранения постоянной функциональности при повреждающих воздействиях [96–98, 146, 147]. Участие ПГД в регенерации связано с большей устойчивостью полиплоидных клеток по сравнению с диплоидными, что особенно необходимо при восстановлении. Например, при необходимости быстрого восстановления функции полиплоидия дает возможность увеличить размер клетки (хотя часто и непропорционально числу геномов), минуя энергетически затратный митоз, который связан с реорганизацией цитоскелета, нарушением межклеточных контактов и архитектуры ткани [120]. Полиплоидия позволяет также продлить жизнь клеток благодаря повышенной устойчивости к геномному стрессу и апоптотическим сигналам [27]. Недавние полногеномные и экспериментальные исследования показали, что полиплоидия связана с переключением метаболизма на режим экономии энергии [93, 94]. Это наглядно проявляется в сердце, где полиплоидия, вызванная стрессом или гиперфункцией, приводит к замещению тяжелой цепи α-миозина (быстрой, взрослой и АТР-затратной) на тяжелую цепь β-миозина (медленную, эмбриональную и АТР-экономную) [100, 102, 103]. Эта взаимосвязь подтверждена на экспериментальных моделях заболеваний сердца, а также гипертонической болезни, дилатационной кардиомиопатии, инфаркте миокарда и ишемии [99, 125].
Способность полиплоидных клеток поддерживать функцию в условиях дефицита энергии и гипоксии подтверждают примеры из дикой природы. Например, около 80% ядер кардиомиоцитов голого землекопа (Heterocephalus glaber) содержат четыре или более геномов [148]. Примечательно, что этот грызун, который обитает в токсических условиях при пониженной концентрации кислорода и живет 32 года, что примерно в 10 раз дольше, чем мышь сходного веса [132]. Известно, что кардиомиоциты других грызунов сходного веса содержат практически только диплоидные ядра [93, 149].
Важно отметить, что ПГД играет особенную роль в регенерации сердца человека, где скорость обновления кардиомиоцитов крайне низка. Недавно показали, что у пожилых людей (в возрасте около 75 лет) в миокарде за год обновляется всего около 0.4% кардиомиоцитов и около 55% кардиомиоцитов представлено клетками, сохранившимися с рождения [96]. В то же время, в миокарде человека преобладают полиплоидные кардиомиоциты. Например, в ядре практически каждого кардиомиоцита (около 90% клеток) здорового человека содержится 4–16 геномов [96, 97]. При патологической гиперфункции кардиомиоциты накапливают по 32, 64 или даже 128 геномов [97, 98]. Таким образом, ПГД является важным альтернативным способом восстановления ткани сердца [98]. Важно отметить, что активация ПГД в кардиомиоцитах патологически измененного сердца может быть в некоторых аспектах даже лучше, чем индукция митоза. С одной стороны, дополнительные ПГД могут улучшать сократительную функцию сердца в условиях гипоксического и механического стресса [97, 98], а с другой, дают возможность избежать побочных эффектов индукции митоза в виде массового апоптоза из-за нарушенной функции центриолей [98].
Полиплоидия может быть механизмом регенерации ткани при старении и заживлении ран, когда пролиферативного потенциала диплоидных клеток не хватает для восстановления дефектов ДНК, цитоскелета, и других компонентов клетки [2, 68]. Важная роль полиплоидии показана при восстановлении функции почек и сердца после ранения и инфаркта [120]. Недавно показали, что эпителиальные клетки почечных канальцев накапливают геномы в ответ на острое повреждение почек, а рост числа полиплоидных клеток помогает восстановить функцию органа [120]. В целом, можно сказать, что регенерация с помощью полиплоидии обеспечивает запас прочности организма при невозможности нормальной регенерации за счет пролиферации диплоидных клеток.
ПРИКЛАДНОЕ ЗНАЧЕНИЕ ПОЛИПЛОИДИИ
Полиплоидия в биотехнологии сельского хозяйства и аквакультуры
Полиплоидия – один из ключевых факторов при одомашнивании сельскохозяйственных культур [23, 46, 150]. Случаев полиплоидии в истории одомашненных растений было больше, чем у их диких предшественников [150]. Генетическая пластичность множественных генов полиплоидного генома представляет особое преимущество для одомашнивания [62]. В число недавних полиплоидов входят такие основные культуры, как пшеница, овес, кукуруза, хлопок, картофель, бобовые, банан, сахарный тростник, рапс, клубника, кофе, горчица, табак. В основном это аллополиплоиды (т.е. результат полиплоидизации, связанной с гибридизацией). Гибридизация часто сопровождается повышенной гетерозиготностью и гетерозисом, тогда как полиплоидизация восстанавливает фертильность образовавшихся гибридов и способствует стабилизации гибридного генома [23].
Аллополиплоидизация открывает путь к созданию новых культур. Синтетические полиплоиды используются для улучшения агрономических признаков, поскольку обладают более высокой приспособленностью, устойчивостью к болезням, быстрым ростом и высокой продуктивностью по сравнению с их природными диплоидами [151, 152]. Так, например, синтетическая гексаплоидная пшеница представляет новый источник генетического разнообразия для получения растений, устойчивых к множественному биотическому стрессу [88]. Многие искусственные полиплоиды используются в аквакультуре и большинство из них созданы из природных полиплоидных моллюсков и рыб, особенно карповых и лососевых [22]. Между родительскими субгеномами в полиплоидных гибридах возникает конкуренция, называемая “доминированием субгенома”, когда менее экспрессированный субгеном имеет тенденцию терять больше генов и цис-регуляторных элементов по сравнению с другим субгеномом [23, 46]. В крайних случаях родительские субгеномы могут даже сегрегировать отдельно в линии зародышевых клеток, и гибридное состояние возникает de novo в каждом поколении [153, 154]. Такое рекурсивное воспроизводство полиплоидных гибридов используется, когда аллополиплоиды стерильны или утрачивают свои гибридные свойства из-за инбридинга. Например, аллогексаплоидный карп массово воспроизводится путем скрещивания родительских видов в каждом поколении [22].
ЗАКЛЮЧЕНИЕ
Полиплоидия широко распространена на уровне целых организмов и в отдельных клетках тканей многоклеточных. Полиплоидия организмов является важной движущей силой эволюции. Увеличивая размах генетической изменчивости, полиплоидия повышает толерантность к широкому спектру стрессовых условий, способствует освоению новых экологических ниш и маскирует вредные последствия мутаций. Соматическая полиплоидия в той или иной степени присутствует во всех тканях многоклеточных организмов. У человека полиплоидные клетки образуются в ходе нормального органогенеза и считаются проявлением программ развития. Накопление геномов может также быть реакцией на стресс, проявлением канцерогенеза, старения и воспаления. Обширные сравнительные исследования показали, что полиплоидия, с одной стороны, усиливает адаптацию к выживанию в экстремальных условиях (особенно при гипоксии), а с другой стороны, ассоциирована со снижением функционального потенциала органов (сердца и печени). В генотоксических условиях, когда вхождение клетки в митоз неминуемо ведет к гибели, полиплоидия служит адаптивной заменой завершенного клеточного цикла. В зависимости от митотического потенциала клеток и тканей полиплоидия может быть опасной или полезной. С одной стороны, удвоение геномов – это эволюционно консервативный способ регенерации и обновления тканей, с другой, полиплоидия – это первый шаг к возникновению опухолеродных клеток и клеток, устойчивых к цитостатикам. Полнотранскриптомные сравнительные исследования показали, что полиплоидия организмов, а также соматических и раковых клеток ассоциирована с рядом общих признаков, которые позволяют понять, какие именно изменения в экспрессии приводят к возникновению адаптивного фенотипа. Это может быть как усилением эволюционно древних программ одноклеточности, которые повышают устойчивость к стрессу, так и активацией путей морфогенеза, которые повышают биологическую пластичность и способствуют адаптивной перестройке обмена. Таким образом, в нашем обзоре показаны общие проявления ПГД на разном уровне организации, в разном эволюционном масштабе и в разных областях исследований. Интеграция данных эволюционной и молекулярной биологии, биохимии, цитологии и молекулярной медицины дала возможность идентифицировать набор эволюционно консервативных признаков ПГД, одновременно поддержанных отбором, которые целесообразно изучать в комплексе. Представленные сведения, улучшают понимание роли полиплоидии в эволюции, органогенезе, старении и канцерогенезе, что может способствовать разработке новых стратегий управления органогенезом и регенерацией, а также предотвращения трансформации клеток.
Авторы признательны Екатерине Эренпрейсе (Латвийский центр биомедицинских исследований и обучения), А.С. Цимохе, Е.В. Чихиржиной и И.Е. Негановой (ИНЦ РАН) за важные комментарии. Мы также признательны анонимным рецензентам за ценные рекомендации и исправления, которые улучшили обзор.
Работа выполнена при финансовой поддержке Российского фонда фундаментальных исследований (программа “Экспансия”, грант № 19-14-50401).
Настоящая статья не содержит каких-либо исследований с участием людей или животных в качестве объектов исследований.
Авторы заявляют об отсутствии конфликта интересов.
Список литературы
Van de Peer Y., Mizrachi E., Marchal K. (2017) The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424.
Fox D.T., Soltis D.E., Soltis P.S., Ashman T.L., Van de Peer Y. (2020) Polyploidy: a biological force from cells to ecosystems. Trends Cell Biol. 30, 688–694.
Ohno S. (1970) Evolution by Gene Duplication. Berlin, Heidelberg: Springer.
Singh P.P., Isambert H. (2020) OHNOLOGS v2: a comprehensive resource for the genes retained from whole genome duplication in vertebrates. Nucl. Acids Res. 48, D724–730.
Kreiner J.M., Kron P., Husband B.C. (2017) Frequency and maintenance of unreduced gametes in natural plant populations: associations with reproductive mode, life history and genome size. New Phytol. 214, 879–889.
Clark J.W., Donoghue P.C.J. (2017) Constraining the timing of whole genome duplication in plant evolutionary history. Proc. Biol. Sci. 284, 20170912.
Murat F., Armero A., Pont C., Klopp C., Salse J. (2017) Reconstructing the genome of the most recent common ancestor of flowering plants. Nat. Genet. 49, 490–496.
Soppa J. (2017) Polyploidy and community structure. Nat. Microbiol. 2, 16261.
Hu G., Wendel J.F. (2019) Cis-trans controls and regulatory novelty accompanying allopolyploidization. New Phytol. 221, 1691–1700.
Rice A., Šmarda P., Novosolov M., Drori M., Glick L., Sabath N., Meiri S., Belmaker J., Mayrose I. (2019) The global biogeography of polyploid plants. Nat. Ecol. Evol. 3, 265–273.
Yang L. Sado T., Hirt M.V., Pasco-Viel E., Arunachalam M., Li J.B., Wang X.Z., Freyhof J., Saitoh K., Simons A.M., Miya M., He S.P., Mayden R.L. (2015) Phylogeny and polyploidy: resolving the classification of cyprinine fishes (Teleostei: Cypriniformes). Mol. Phylogenet. Evol. 85, 97–116.
Evans B.J., Carter T.F., Greenbaum E., Gvoždík V., Kelley D.B., McLaughlin P.J., Olivier Pauwels S.G., Portik D.M., Stanley E.L., Tinsley R.C., Tobias M.L., Blackburn D.C. (2015) Genetics, morphology, advertisement calls, and historical records distinguish six new polyploid species of african clawed frog (Xenopus, Pipidae) from West and Central Africa. PLoS One. 10, e0142823.
Moritz C., Bi K. (2011) Spontaneous speciation by ploidy elevation: laboratory synthesis of a new clonal vertebrate. Proc. Natl. Acad. Sci. USA. 108, 9733–9734.
Tiersch T.R., Beck M.L., Douglass M. (1991) ZZW autotriploidy in a blue-and-yellow macaw. Genetica. 84, 209–212.
Evans B.J., Upham N.S., Golding G.B., Ojeda R.A., Ojeda A.A. (2017) Evolution of the largest mammalian genome. Genome Biol. Evol. 9, 1711–1724.
Imai H., Fujii W., Kusakabe K.T., Kiso Y., Kano K. (2016) Effects of whole genome duplication on cell size and gene expression in mouse embryonic stem cells. J. Reprod. Dev. 62, 571–576.
Otto S.P., Whitton J. (2000) Polyploid incidence and evolution. Annu. Rev. Genet. 34, 401–437.
Ganem N.J., Pellman D. (2007) Limiting the proliferation of polyploid cells. Cell. 131, 437–440.
Otto S.P. (2007) The evolutionary consequences of polyploidy. Cell. 131, 452–462.
Cutie S., Huang G.N. (2021) Vertebrate cardiac regeneration: evolutionary and developmental perspectives. Cell Regen. 10, 6.
Reyes A.A., Marcum R.D., He Y. (2021) Structure and function of chromatin remodelers. J. Mol. Biol. 433, 166929.
Zhou L., Gui J. (2017) Natural and artificial polyploids in aquaculture. Aquaculture and Fisheries. 2, 103–111.
Glombik M., Bačovský V., Hobza R., Kopecký D. (2020) Competition of parental genomes in plant hybrids. Front. Plant. Sci. 11, 200.
Comai L. (2005) The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846.
Anatskaya O.V., Vinogradov A.E., Kudryavtsev B.N. (1994) Hepatocyte polyploidy and metabolism/life-history traits: hypotheses testing. J. Theor. Biol. 168, 191–199.
Gemble S., Basto R. (2020) CHRONOCRISIS: when cell cycle asynchrony generates DNA damage in polyploid cells. Bioessays. 42, e2000105.
Øvrebø J.I., Edgar B.A. (2018) Polyploidy in tissue homeostasis and regeneration. Development. 145, dev156034.
Brukman N.G., Uygur B., Podbilewicz B., Chernomordik L.V. (2019) How cells fuse. J. Cell Biol. 218, 1436–1451.
Gjelsvik K.J., Besen-McNally R., Losick V.P. (2019) Solving the polyploid mystery in health and disease. Trends Genet. 35, 6–14.
Anatskaya O.V., Vinogradov A.E. (2007) Genome multiplication as adaptation to tissue survival: evidence from gene expression in mammalian heart and liver. Genomics. 89, 70–80.
Schoenfelder K.P., Fox D.T. (2015) The expanding implications of polyploidy. J. Cell Biol. 209, 485–491.
Varetti G., Pellman D. (2012) “Two” much of a good thing: telomere damage-induced genome doubling drives tumorigenesis. Cancer Cell. 21, 712–714.
Priestley P., Baber J., Lolkema M.P., Steeghs N., de Bruijn E., Shale C., Duyvesteyn K., Haidari S., van Hoeck A., Onstenk W., Roepman P., Voda M., Bloemendal H.J., Tjan-Heijnen V.C.G., van Herpen C.M.L., Labots M., Witteveen P.O., Smit E.F., Sleijfer S., Voest E.E., Cuppen E. (2019) Pan-cancer whole-genome analyses of metastatic solid tumours. Nature. 575, 210–216.
White-Gilbertson S., Voelkel-Johnson C. (2020) Giants and monsters: unexpected characters in the story of cancer recurrence. Adv. Cancer Res. 148, 201–232.
Salmina K., Bojko A., Inashkina I., Staniak K., Dudkowska M., Podlesniy P., Rumnieks F., Vainshelbaum N.M., Pjanova D., Sikora E., Erenpreisa J. (2020) “Mitotic slippage” and extranuclear DNA in cancer chemoresistance: a focus on telomeres. Int. J. Mol. Sci. 21, 2779.
Salmina K., Jankevics E., Huna A., Perminov D., Radovica I, Klymenko T., Ivanov A., Jascenko E., Scherthan H., Cragg M., Erenpreisa J. (2010) Up-regulation of the embryonic self-renewal network through reversible polyploidy in irradiated p53-mutant tumour cells. Exp. Cell Res. 316, 2099–2112.
Erenpreisa J., Kalejs M., Cragg M.S. (2005) Mitotic catastrophe and endomitosis in tumour cells: an evolutionary key to a molecular solution. Cell Biol. Int. 29, 1012–1018.
Walen K.H. (2012) Genome reversion process of endopolyploidy confers chromosome instability on the descendent diploid cells. Cell Biol. Int. 36, 137–145.
Walen K.H. (2021) Cell cycle stress in normal human cells: a route to “first cells” (with/without fitness gain) and cancer-like cell-shape changes. Semin. Cancer Biol. https://doi.org/10.1016/j.semcancer.2020.12.023
Cho Y., Kim Y.K. (2020) Cancer stem cells as a potential target to overcome multidrug resistance. Front Oncol. 10, 764.
Liu J. (2018) The dualistic origin of human tumors. Semin. Cancer Biol. 53, 1–16.
Erenpreisa J., Salmina K., Anatskaya O., Cragg M.S. (2020) Paradoxes of cancer: survival at the brink. Semin Cancer Biol. S1044-579X(20)30269-8. https://doi.org/10.1016/j.semcancer.2020.12.009
Du K., Klopp C., Woltering J.M., Adolfi M.C., Feron R., Prokopov D., Makunin A., Kichigin I., Schmidt C., Fischer P., Kuhl H., Wuertz S., Gessner J., Kloas W., Cabau C., Iampietro C., Parrinello H., Tomlinson C., Journot L., Postlethwait J.H., Braasch I., Trifonov V., Warren W.C., Meyer A., Guiguen Y., Schartl M. (2020) The sterlet sturgeon genome sequence and the mechanisms of segmental rediploidization. Nat. Ecol. Evol. 4, 841–852.
Robertson F.M., Gundappa M.K., Grammes F., Hvidsten T.R., Redmond A.K., Lien S., Martin S.A.M., Holland P.W.H., Sandve S.R., Macqueen D.J. (2017) Lineage-specific rediploidization is a mechanism to explain time-lags between genome duplication and evolutionary diversification. Genome Biol. 18, 111.
Venkatachalam A.B., Parmar M.B., Wright J.M. (2017) Evolution of the duplicated intracellular lipid-binding protein genes of teleost fishes. Mol. Genet. Genomics. 292, 699–727.
Cheng F., Wu J., Cai X., Liang J., Freeling M., Wang X. (2018) Gene retention, fractionation and subgenome differences in polyploid plants. Nat. Plants. 4, 258–268.
Lynch M., Force A. (2000) The probability of duplicate gene preservation by subfunctionalization. Genetics. 154, 459–473.
Conant G.C., Wolfe K.H. (2008) Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950.
Vinogradov A.E. (2009) Global versus local centrality in evolution of yeast protein network. J. Mol. Evol. 68, 192–196.
Vinogradov A.E., Anatskaya O.V. (2009) Loss of protein interactions and regulatory divergence in yeast whole-genome duplicates. Genomics. 93, 534–542.
Lucchetta E.M., Ohlstein B. (2017) Amitosis of polyploid cells regenerates functional stem cells in the Drosophila intestine. Cell Stem Cell. 20, 609–620.e6.
Zybina T.G., Zybina E.V. (2020) Role of cell cycling and polyploidy in placental trophoblast of different mammalian species. Reprod. Domest. Anim. 55, 895–904.
Ahmadbeigi N., Soleimani M., Vasei M., Gheisari Y., Mortazavi Y., Azadmanesh K., Omidkhoda A., Janzamin E., Nardi N.B. (2013) Isolation, characterization, and transplantation of bone marrow-derived cell components with hematopoietic stem cell niche properties. Stem Cells Dev. 22, 3052–3061.
Erenpreisa J., Cragg M.S. (2010) MOS, aneuploidy and the ploidy cycle of cancer cells. Oncogene. 29, 5447–5451.
Niu N., Mercado-Uribe I., Liu J. (2017) Dedifferentiation into blastomere-like cancer stem cells via formation of polyploid giant cancer cells. Oncogene. 36, 4887–4900.
Niculescu V.F. (2019) The reproductive life cycle of cancer: hypotheses of cell of origin, TP53 drivers and stem cell conversions in the light of the atavistic cancer cell theory. Med. Hypotheses. 123, 19–23.
Liu J. (2020) The “life code”: A theory that unifies the human life cycle and the origin of human tumors. Semin. Cancer Biol. 60, 380–397.
Niculescu V.F. (2020) aCLS cancers: genomic and epigenetic changes transform the cell of origin of cancer into a tumorigenic pathogen of unicellular organization and lifestyle. Gene. 726, 144174.
Corrochano L.M., Kuo A., Marcet-Houben M., Polaino S., Salamov A., Villalobos-Escobedo J.M., Grimwood J., Álvarez M.I., Avalos J., Bauer D., Benito E.P., Benoit I., Burger G., Camino L.P., Cánovas D., Cerdá-Olmedo E., Cheng J.F., Domínguez A., Eliáš M., Eslava A.P., Glaser F., Gutiérrez G., Heitman J., Henrissat B., Iturriaga E.A., Lang B.F., Lavín J.L., Lee S.C., Li W., Lindquist E., López-García S., Luque E.M., Marcos A.T., Martin J., McCluskey K., Medina H.R., Miralles-Durán A., Miyazaki A., Muñoz-Torres E., Oguiza J.A., Ohm R.A., Olmedo M., Orejas M., Ortiz-Castellanos L., Pisabarro A.G., Rodríguez-Romero J., Ruiz-Herrera J., Ruiz-Vázquez R., Sanz C., Schackwitz W., Shahriari M., Shelest E., Silva-Franco F., Soanes D., Syed K., Tagua V.G., Talbot N.J., Thon M.R., Tice H., de Vries R.P., Wiebenga A., Yadav J.S., Braun E.L., Baker S.E., Garre V., Schmutz J., Horwitz B.A., Torres-Martínez S., Idnurm A., Herrera-Estrella A., Gabaldón T., Grigoriev I.V. (2016) Expansion of signal transduction pathways in fungi by extensive genome duplication. Curr. Biol. 26, 1577–1584.
Vinogradov A.E., Anatskaya O.V. (2019) Evolutionary framework of the human interactome: unicellular and multicellular giant clusters. Biosystems. 181, 82–87.
Mattenberger F., Sabater-Muñoz B., Toft C., Fares M.A. (2017) The phenotypic plasticity of duplicated genes in Saccharomyces cerevisiae and the origin of adaptations. G3 (Bethesda). 7, 63–75.
Zhang K., Wang X., Cheng F. (2019) Plant polyploidy: origin, evolution, and its influence on crop domestication. Horticult. Plant J. 5, 231–239.
Katsuda T., Hosaka K., Matsuzaki J., Usuba W., Prieto-Vila M., Yamaguchi T., Tsuchiya A., Terai S., Ochiya T. (2020) Transcriptomic dissection of hepatocyte heterogeneity: linking ploidy, zonation, and stem/progenitor cell characteristics. Cell. Mol. Gastroenterol. Hepatol. 9, 161–183.
Broughton K.M., Khieu T., Nguyen N., Rosa M., Mohsin S., Quijada P., Wang B.J., Echeagaray O.H., Kubli D.A., Kim T., Firouzi F., Monsanto M.M., Gude N.A., Adamson R.M., Dembitsky W.P., Da-vis M.E., Sussman M.A. (2019) Cardiac interstitial tetraploid cells can escape replicative senescence in rodents but not large mammals. Commun. Biol. 2, 205.
Fajka-Boja R., Marton A., Tóth A., Blazsó P., Tubak V., Bálint B., Nagy I., Hegedűs Z., Vizler C., Katona R.L. (2018) Increased insulin-like growth factor 1 production by polyploid adipose stem cells promotes growth of breast cancer cells. BMC Cancer. 18, 872.
Anatskaya O.V., Vinogradov A.E., Vainshelbaum N.M., Giuliani A., Erenpreisa J. (2020) Phylostratic shift of whole-genome duplications in normal mammalian tissues towards unicellularity is driven by developmental bivalent genes and reveals a link to cancer. Int. J. Mol. Sci. 21, 8759.
Vinogradov A.E., Anatskaya O.V. (2020) Cell-cycle dependence of transcriptome gene modules: comparison of regression lines. FEBS J. 287, 4427–4439.
Moein S., Adibi R., da Silva Meirelles L., Nardi N.B., Gheisari Y. (2020) Cancer regeneration: polyploid cells are the key drivers of tumor progression. Biochim. Biophys. Acta – Rev. Cancer. 1874, 188408.
Krigerts J., Salmina K., Freivalds T., Zayakin P., Rumnieks F., Inashkina I., Giuliani A., Hausmann M., Erenpreisa J. (2021) Differentiating cancer cells reveal early large-scale genome regulation by pericentric domains. Biophys. J. 120, 711–724.
Pienta K.J., Hammarlund E.U., Brown J.S., Amend S.R., Axelrod R.M. (2021) Cancer recurrence and lethality are enabled by enhanced survival and reversible cell cycle arrest of polyaneuploid cells. Proc. Natl. Acad. Sci. USA. 118, e2020838118.
Neganova I., Zhang X., Atkinson S., Lako M. (2009) Expression and functional analysis of G1 to S regulatory components reveals an important role for CDK2 in cell cycle regulation in human embryonic stem cells. Oncogene. 28, 20–30.
Neganova I., Vilella F., Atkinson S.P., Lloret M., Passos J.F., von Zglinicki T., O’Connor J.E., Burks D., Jones R., Armstrong L., Lako M. (2011) An important role for CDK2 in G1 to S checkpoint activation and DNA damage response in human embryonic stem cells. Stem Cells. 29, 651–659.
Neganova I., Tilgner K., Buskin A., Paraskevopoulou I., Atkinson S.P., Peberdy D., Passos J.F., Lako M. (2014) CDK1 plays an important role in the maintenance of pluripotency and genomic stability in human pluripotent stem cells. Cell Death Dis. 5, e1508.
Vinogradov A.E., Shilina M.A., Anatskaya O.V., Alekseenko L.L., Fridlyanskaya I.I., Krasnenko A., Kim A., Korostin D., Ilynsky V., Elmuratov A., Tsy-ganov O., Grinchuk T.M., Nikolsky NN. (2017) Molecular genetic analysis of human endometrial mesenchymal stem cells that survived sublethal heat shock. Stem Cells Int. 2017, 2362630.
Shilina M.A., Grinchuk T.M., Anatskaya O.V., Vinogradov A.E., Alekseenko L.L., Elmuratov A.U., Nikolsky N.N. (2018) Cytogenetic and transcriptomic analysis of human endometrial MSC retaining proliferative activity after sublethal heat shock. Cells. 7, 184.
Alekseenko L.L., Shilina M.A., Lyublinskaya O.G., Kornienko J.S., Anatskaya O.V., Vinogradov A.E., Grinchuk T.M., Fridlyanskaya I.I., Nikolsky N.N. (2018) Quiescent human mesenchymal stem cells are more resistant to heat stress than cycling cells. Stem Cells Int. 2018, 3753547.
Vincent M.D. (2009) Optimizing the management of advanced non-small-cell lung cancer: a personal view. Curr. Oncol. 16, 9–21.
Davies P.C.W., Lineweaver C.H. (2011) Cancer tumors as Metazoa 1.0: tapping genes of ancient ancestors. Phys. Biol. 8, 015001.
Vinogradov A.E. (2010) Human transcriptome nexuses: basic-eukaryotic and metazoan. Genomics. 95, 345–354.
Trigos A.S., Pearson R.B., Papenfuss A.T., Goode D.L. (2017) Altered interactions between unicellular and multicellular genes drive hallmarks of transformation in a diverse range of solid tumors. Proc. Natl. Acad. Sci. USA. 114, 6406–6411.
Erenpreisa J., Salmina K., Huna A., Jackson T.R., Vazquez-Martin A., Cragg M.S. (2014) The “virgin birth”, polyploidy, and the origin of cancer. Oncoscience. 2, 3–14.
Matsumoto T., Wakefield L., Peters A., Peto M., Spellman P., Grompe M. (2021) Proliferative polyploid cells give rise to tumors via ploidy reduction. Nat. Commun. 12, 646.
Zhang S., Mercado-Uribe I., Xing Z., Sun B., Kuang J., Liu J. (2014) Generation of cancer stem-like cells through the formation of polyploid giant cancer cells. Oncogene. 33, 116–128.
Zhang S., Mercado-Uribe I., Sood A., Bast R.C., Liu J. (2016) Coevolution of neoplastic epithelial cells and multilineage stroma via polyploid giant cells during immortalization and transformation of mullerian epithelial cells. Genes Cancer. 7, 60–72.
Kozlov A.P. (2014) Evolution by Tumor Neofunctionalization. Elsevier/Acad. Press. 248 p.
Kozlov A.P. (2019) The role of heritable tumors in evolution of development: a new theory of carcino-evo-devo. Acta Naturae. 11, 65–72.
Ruiz M., Quiñones A., Martínez-Cuenca M.R., Aleza P., Morillon R., Navarro L., Primo-Millo E., Martínez-Alcántara B. (2016) Tetraploidy enhances the ability to exclude chloride from leaves in Carrizo citrange seedlings. J. Plant Physiol. 205, 1–10.
Bhatta M., Morgounov A., Belamkar V., Wegulo S.N., Dababat A.A., Erginbas-Orakci G., Bouhssini M.E., Gautam P., Poland J., Akci N., Demir L., Wanyera R., Baenziger P.S. (2019) Genome-wide association study for multiple biotic stress resistance in synthetic hexaploid wheat. Int. J. Mol. Sci. 20, 3667.
Yao Y., Carretero-Paulet L., Van de Peer Y. (2019) Using digital organisms to study the evolutionary consequences of whole genome duplication and polyploidy. PLoS One. 14, e0220257.
Keane O.M., Toft C., Carretero-Paulet L., Jones G.W., Fares M.A. (2014) Preservation of genetic and regulatory robustness in ancient gene duplicates of Saccharomyces cerevisiae. Genome Res. 24, 1830–1841.
Carretero-Paulet L., Van de Peer Y. (2020) The evolutionary conundrum of whole-genome duplication. Am. J. Bot. 107, 1101–1105.
Анацкая О.В., Виноградов А.Е., Кудрявцев Б.Н. (1998) Уровни плоидности миоцитов в разных отделах сердца птиц. Цитология. 5, 359–371.
Anatskaya O.V., Vinogradov A.E. (2004) Paradoxical relationship between protein content and nucleolar activity in mammalian cardiomyocytes. Genome. 47, 565–578.
Anatskaya O.V., Vinogradov A.E. (2004) Heart and liver as developmental bottlenecks of mammal design: evidence from cell polyploidization. Biol. J. Linn. Soc. 83, 175–186.
Anatskaya O.V., Vinogradov A.E. (2002) Myocyte ploidy in heart chambers of birds with different locomotor activity. J. Exp. Zool. 293, 427–441.
Derks W., Bergmann O. (2020) Polyploidy in cardiomyocytes: roadblock to heart regeneration? Circ. Res. 126, 552–565.
Brodsky V.Y., Sarkisov D.S., Arefyeva A.M., Panova N.W., Gvasava I.G. (1994) Polyploidy in cardiac myocytes of normal and hypertrophic human hearts; range of values. Virchows Arch. 424, 429–435.
Leone M., Engel F.B. (2019) Advances in heart regeneration based on cardiomyocyte proliferation and regenerative potential of binucleated cardiomyocytes and polyploidization. Clin. Sci. (Lond). 133, 1229–1253.
Anatskaya O.V., Sidorenko N.V., Beyer T.V., Vinogradov A.E. (2010) Neonatal cardiomyocyte ploidy reveals critical windows of heart development. Int. J. Cardiol. 141, 81–91.
Анацкая О.В., Сидоренко Н.В., Бейер Т.В., Виноградов А.Е. (2010) Неонатальный гастроэнтерит как причина долговременной атрофии, деформации и необратимой гиперполиплоидизации кардиомиоцитов. Кардиология. 10(12), 35–44.
Anatskaya O.V., Sidorenko N.V., Vinogradov A.E., Beyer T.V. (2007) Impact of neonatal cryptosporidial gastroenteritis on epigenetic programming of rat hepatocytes. Cell Biol. Int. 31, 420–427.
Anatskaya O.V., Sidorenko N.V., Matveev I.V., Kropotov A.V., Vinogradov A.E. (2012) Remodeling of rat cardiomyocytes after neonatal cryptosporidiosis. II. Deformation, excessive polyploidization, and HIF-1α overexpression. Cell Tiss. Biol. 6, 472–484.
Anatskaya O.V., Matveev I.V., Sidorenko N.V., Kharchenko M.V., Kropotov A.V., Vinogradov A.E. (2013) Changes in the heart of neonatal rats after cryptosporidial gastroenteritis of different degrees of severity. J. Evol. Biochem. Phys. 49, 509–518.
Bensley J.G., Stacy V.K., De Matteo R., Harding R., Black M.J. (2010) Cardiac remodelling as a result of pre-term birth: implications for future cardiovascular disease. Eur. Heart J. 31, 2058–2066.
Filatova N.A., Knyazev N.A., Skarlato S.O., Anatskaya O.V., Vinogradov A.E. (2018) Natural killer cell activity irreversibly decreases after Cryptosporidium gastroenteritis in neonatal mice. Parasite Immunol. 40, e12524.
Mayfield-Jones D., Washburn J.D., Arias T., Edger P.P., Pires J.C., Conant G.C. (2013) Watching the grin fade: tracing the effects of polyploidy on different evolutionary time scales. Semin. Cell Dev. Biol. 24, 320–331.
Vinogradov A.E., Anatskaya O.V., Kudryavtsev B.N. (2001) Relationship of hepatocyte ploidy levels with body size and growth rate in mammals. Genome. 44, 350–360.
Anatskaya O.V., Vinogradov A.E., Kudryavtsev B.N. (2001) Cardiomyocyte ploidy levels in birds with different growth rates. J. Exp. Zool. 289, 48–58.
Анацкая О.В., Эренпрейса Е.А., Никольский Н.Н., Виноградов А.Е. (2015) Попарно-перекрестное сравнение транскриптомов млекопитающих в исследовании влияния полиплоидии на активность экспрессии генных модулей развития. Цитология. 57, 899–908.
Vinogradov A.E. (2005) Genome size and chromatin condensation in vertebrates. Chromosoma. 113, 362–369.
Pienta K.J., Hammarlund E.U., Axelrod R., Amend S.R., Brown J.S. (2020) Convergent evolution, evolving evolvability, and the origins of lethal cancer. Mol. Cancer Res. 18, 801–810.
Lopez-Sánchez L.M., Jimenez C., Valverde A., Hernandez V., Peñarando J., Martinez A., Lopez-Pedrera C., Muñoz-Castañeda J.R., De la Haba-Rodríguez J.R., Aranda E., Rodriguez-Ariza A. (2014) CoCl2, a mimic of hypoxia, induces formation of polyploid giant cells with stem characteristics in colon cancer. PLoS One. 9, e99143.
Mirzayans R., Murray D. (2020) Intratumor heterogeneity and therapy resistance: contributions of dormancy, apoptosis reversal (Anastasis) and cell fusion to disease recurrence. IJMS. 21, 1308.
Mirzayans R., Andrais B., Murray D. (2018) Roles of polyploid/multinucleated giant cancer cells in metastasis and disease relapse following anticancer treatment. Cancers (Basel). 10, 118.
Amend S.R., Torga G., Lin K.-C., Kostecka L.G., de Marzo A., Austin R.H., Pienta K.J. (2019) Polyploid giant cancer cells: unrecognized actuators of tumorigenesis, metastasis, and resistance. Prostate. 79, 1489–1497.
Anatskaya O.V., Vinogradov A.E. (2010) Somatic polyploidy promotes cell function under stress and energy depletion: evidence from tissue-specific mammal transcriptome. Funct. Integr. Genomics. 10, 433–446.
Vazquez-Martin A., Anatskaya O.V., Giuliani A., Erenpreisa J., Huang S., Salmina K., Inashkina I., Huna A., Nikolsky N.N., Vinogradov A.E. (2016) Somatic polyploidy is associated with the upregulation of c-MYC interacting genes and EMT-like signature. Oncotarget. 7, 75235–75260.
Anatskaya O.V., Erenpreisa J., Giuliani A., Tsimok-ha A.S., Salmina K., Vinogradov A.E. (2020) Polyploidy related induction of morphogenetic signaling is mediated via proteasome pathway. Cell Death Discov. 6, RPC02.
Cеленина А.В., Цимоха А.С., Томилин А.Н. (2017) Протеасомы в регуляции белкового гомеостаза плюрипотентных стволовых клеток. Acta Naturae. 9, 39–47.
Lazzeri E., Angelotti M.L., Conte C., Anders H.-J., Romagnani P. (2019) Surviving acute organ failure: cell polyploidization and progenitor proliferation. Trends Mol. Med. 25, 366–381.
Donne R., Saroul-Aïnama M., Cordier P., Celton-Morizur S., Desdouets C. (2020) Polyploidy in liver development, homeostasis and disease. Nat. Rev. Gastroenterol. Hepatol. 17, 391–405.
Chikhirzhina E., Starkova T., Polyanichko A. (2018) The role of linker histones in chromatin structural organization. 1. H1 family histones. Biophysics. 63, 858–865.
Chikhirzhina E.V., Starkova T.Yu., Polyanichko A.M. (2020) The role of linker histones in chromatin structural organization. 2. Interaction with DNA and nuclear proteins. Biophysics. 65, 202–212.
Chikhirzhina E., Starkova T., Beljajev A., Polyanichko A., Tomilin A. (2020) Functional diversity of non-histone chromosomal protein HmgB1. Int. J. Mol. Sci. 21, 7948.
Старкова Т.Ю., Артамонова Т.О., Ермакова В.В., Чихиржина Е.В., Ходорковский М.А., Томилин А.Н. (2019) Профиль посттранскрипционных модификаций гистона Н1 в хроматине эмбриональных ставоловых клеток мыши. Acta Naturae.11, 82–91.
Gilsbach R., Preissl S., Grüning B.A., Schnick T., Burger L., Benes V., Würch A., Bönisch U., Günther S., Backofen R., Fleischmann B.K., Schübeler D., Hein L. (2014) Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat. Commun. 5, 5288.
Silva I.S., Ghiraldini F.G., Veronezi G.M.B., Mel-lo M.L.S. (2018) Polyploidy and nuclear phenotype characteristics of cardiomyocytes from diabetic adult and normoglycemic aged mice. Acta Histochem. 120, 84–94.
Bian F., Gao F., Kartashov A.V., Jegga A.G., Barski A., Das S.K. (2016) Polycomb repressive complex 1 controls uterine decidualization. Sci. Rep. 6, 26061.
Han P., Li W., Yang J., Shang C., Lin C.H., Cheng W., Hang C.T., Cheng H.L., Chen C.H., Wong J., Xiong Y., Zhao M., Drakos S.G., Ghetti A., Li D.Y., Bernstein D., Chen H.S., Quertermous T., Chang C.P. (2016) Epigenetic response to environmental stress: assembly of BRG1-G9a/GLP-DNMT3 repressive chromatin complex on Myh6 promoter in pathologically stressed hearts. Biochim. Biophys. Acta. 1863, 1772–1781.
Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S.L., Lander E.S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 125, 315–326.
Malik A., Korol A., Weber M., Hankeln T., Avivi A., Band M. (2012) Transcriptome analysis of the spalax hypoxia survival response includes suppression of apoptosis and tight control of angiogenesis. BMC Genomics. 13, 615.
Ma S., Upneja A., Galecki A., Tsai Y.M., Burant C.F., Raskind S., Zhang Q., Zhang Z.D., Seluanov A., Gorbunova V., Clish C.B., Miller R.A., Gladyshev V.N. (2016) Cell culture-based profiling across mammals reveals DNA repair and metabolism as determinants of species longevity. Elife. 5, e19130.
Ma S., Gladyshev V.N. (2017) Molecular signatures of longevity: Insights from cross-species comparative studies. Semin. Cell Dev. Biol. 70, 190–203.
Tsimokha A.S., Kulichkova V.A., Karpova E.V., Zaykova J.J., Aksenov N.D., Vasilishina A.A., Kropotov A.V., Antonov A., Barlev N.A. (2014) DNA damage modulates interactions between microRNAs and the 26S proteasome. Oncotarget. 5, 3555–3567.
Margulis B., Tsimokha A., Zubova S., Guzhova I. (2020) Molecular chaperones and proteolytic machineries regulate protein homeostasis in aging cells. Cells. 9, 1308.
Blanc G., Wolfe K.H. (2004) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 16, 1667–1678.
Fotiou E., Williams S., Martin-Geary A., Robertson D.L., Tenin G., Hentges K.E., Keavney B. (2019) Integration of large-scale genomic data sources with evolutionary history reveals novel genetic loci for congenital heart disease. Circ. Genom. Precis. Med. 12, 442–451.
Yamasaki M., Makino T., Khor S.S., Toyoda H., Miyagawa T., Liu X., Kuwabara H., Kano Y., Shimada T., Sugiyama T., Nishida H., Sugaya N., Tochigi M., Otowa T., Okazaki Y., Kaiya H., Kawamura Y., Miyashita A., Kuwano R., Kasai K., Tanii H., Sasaki T., Honda M., Tokunaga K. (2020) Sensitivity to gene dosage and gene expression affects genes with copy number variants observed among neuropsychiatric diseases. BMC Med. Genomics. 13, 55.
Arbabian A., Iftinca M., Altier C., Singh P.P., Isambert H., Coscoy S. (2020) Mutations in calmodulin-binding domains of TRPV4/6 channels confer invasive properties to colon adenocarcinoma cells. Channels (Austin). 14, 101–109.
Illidge T.M., Cragg M.S., Fringes B., Olive P., Erenpreisa J.A. (2000) Polyploid giant cells provide a survival mechanism for p53 mutant cells after DNA damage. Cell Biol. Int. 24, 621–633.
Sundaram M., Guernsey D.L., Rajaraman M.M., Rajaraman R. (2004) Neosis: a novel type of cell division in cancer. Cancer Biol. Ther. 3, 207–218.
Puig P.-E., Guilly M.N., Bouchot A., Droin N., Cathelin D., Bouyer F., Favier L., Ghiringhelli F., Kroemer G., Solary E., Martin F., Chauffert B. (2008) Tumor cells can escape DNA-damaging cisplatin through DNA endoreduplication and reversible polyploidy. Cell Biol. Int. 32, 1031–1043.
Lagadec C., Vlashi E., Della Donna L., Dekmezian C., Pajonk F. (2012) Radiation-induced reprogramming of breast cancer cells: radiation-induced cancer stem cells. Stem Cells. 30, 833–844.
Weihua Z., Lin Q., Ramoth A.J., Fan D., Fidler I.J. (2011) Formation of solid tumors by a single multinucleated cancer cell. Cancer. 117, 4092–4099.
Sikora E., Czarnecka-Herok J., Bojko A., Sunderland P. (2020) Therapy-induced polyploidization and senescence: coincidence or interconnection? Semin. Cancer Biol. S1044-579X(20)30253-4. https://doi.org/10.1016/j.semcancer.2020.11.015
Patterson M., Swift S.K. (2019) Residual diploidy in polyploid tissues: a cellular state with enhanced proliferative capacity for tissue regeneration? Stem Cells Dev. 28, 1527–1539.
Patterson M., Barske L., Van Handel B., Rau C.D., Gan P., Sharma A., Parikh S., Denholtz M., Huang Y., Yamaguchi Y., Shen H., Allayee H., Crump J.G., Force T.I., Lien C.L., Makita T., Lusis A.J., Kumar S.R., Sucov H.M. (2017) Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nat. Genet. 49, 1346–1353.
Gan P., Patterson M., Velasquez A., Wang K., Tian D., Windle J.J., Tao G., Judge D.P., Makita T., Park T.J., Sucov H.M. (2019) Tnni3k alleles influence ventricular mononuclear diploid cardiomyocyte frequency. PLoS Genet. 15, e1008354.
Анацкая О.В., Рунов А.Л., Вонский М.С., Харченко М.В., Пономарцев С.В., Елмуратов А.У., Виноградов А.Е. (2019) Нарушение постнатального органогенеза сердца после неонатальной непереносимости лактозы. Гены и клетки. 14, 21–22.
Salman-Minkov A., Sabath N., Mayrose I. (2016) Whole-genome duplication as a key factor in crop domestication. Nat. Plants. 2, 16115.
Guo H., Mendrikahy J.N., Xie L., Deng J., Lu Z., Wu J., Li X., Shahid M.Q., Liu X. (2017) Transcriptome analysis of neo-tetraploid rice reveals specific differential gene expressions associated with fertility and heterosis. Sci. Rep. 7, 40139.
Julião S.A., Ribeiro C.D.V., Lopes J.M.L., de Matos E.M., Reis A.C., Peixoto P.H.P., Machado M.A., Azevedo A.L.S., Grazul R.M., de Campos J.M.S., Viccini L.F. (2020) Induction of synthetic polyploids and assessment of genomic stability in Lippia alba. Front. Plant Sci. 11, 292.
Vinogradov A.E., Borkin L.J., Günther R., Rosa-nov J.M. (1991) Two germ cell lineages with genomes of different species in one and the same animal. Hereditas. 114, 245–251.
Vinogradov A.E., Borkin L.J., Günther R., Rosanov J.M. (1990) Genome elimination in diploid and triploid Rana esculenta males: cytological evidence from DNA flow cytometry. Genome. 33, 619–627.
Дополнительные материалы отсутствуют.
Инструменты
Молекулярная биология