Молекулярная биология, 2022, T. 56, № 3, стр. 391-417
Гетерогенность популяции митохондрий в клетках растений и других организмов
Т. А. Тарасенко a, *, М. В. Кулинченко a
a Сибирский институт физиологии и биохимии растений Сибирского отделения Российской академии наук
664033 Иркутск, Россия
* E-mail: bolotova_t.a@mail.ru
Поступила в редакцию 22.09.2021
После доработки 10.10.2021
Принята к публикации 11.10.2021
- EDN: JEPVNT
- DOI: 10.31857/S0026898422020185
Аннотация
Популяция митохондрий в клетках эукариот отличается неоднородностью. Гетерогенность митохондрий может быть определена как вариация тех или иных характеристик митохондрий в пределах одной или разных клеток. Различия между митохондриями могут быть отнесены к негенетическим (структурно-морфологические и биоэнергетические особенности) или к генетическим (различия в числе копий или в последовательности митохондриальной (мт) ДНК). Изменения в последовательности мтДНК могут находить отражение в явлении гетероплазмии, то есть сосуществовании в клетке/организме различных митохондриальных генотипов. В обзоре рассмотрены особенности организации и динамики хондриома клеток растений в сравнении с другими таксономическими группами организмов. Особое внимание уделено причинам и механизмам, ведущим к митохондриальной гетерогенности, феномену гетероплазмии у растений и возможности функциональной специализации у митохондрий, а также роли этих процессов для всего организма. Анализ многочисленных данных показывает, что причиной неоднородного состояния митохондрий в клетке могут быть разные факторы, в том числе видоспецифические особенности процессов митохондриальной динамики, отвечающие за гомогенность популяции этих органелл клетки.
ВВЕДЕНИЕ
В клетках эукариотических организмов митохондрии выполняют важнейшие функции, такие как осуществление энергетического метаболизма клетки, запуск клеточной дифференцировки и инициирование гибели клеток. Известно, что митохондрии в клетке могут различаться своей морфологией (форма, размер, плотность) и структурой (сферические, удлиненные, разветвленные органеллы). Различия в морфологии и структуре хондриома клетки находятся в прямой зависимости от видовой принадлежности и стадии развития эукариотического организма, клеточной и тканевой специализации или метаболического состояния клетки [1]. Структура и функции хондриома зависят от стадии биогенеза органелл, митотической сегрегации, влияющей на локализацию органелл в клетке, и опосредуются консервативным среди всех эукариот механизмом митохондриальной динамики. Этот механизм включает в себя два взаимосвязанных процесса: деления и слияния митохондрий [2], ‒ которые во многом определяют морфологическую пластичность органелл [3]. В соматических клетках растений митохондриальную популяцию чаще всего составляют дискретные органеллы. Их структура поддерживается определенным балансом между процессами деления и слияния, который у растений, по всей видимости, смещен в сторону деления.
Наряду с ядром, хоть и в несопоставимом по объему количестве, митохондрии несут наследственную информацию. Для наследственной информации характерны два взаимодополняющих параметра: стабильность и изменчивость. Баланс стабильности и вариативности цитоплазматической наследственности осуществляется при участии процессов слияния-деления. Благодаря этому, в клетке поддерживается гомогенность митохондриальной популяции и сохраняется стабильность наследования генетической информации, но в то же время и разнообразие органелл. Митохондриальная гетерогенность определяется различиями в составе и числе копий генома, в активности некоторых ферментов, в эффективности транспорта макромолекул [4‒7]. Так, например, процесс импорта белков и нуклеиновых кислот в митохондрии, влияющий на генетическую систему этих органелл и их биогенез, может иметь разную эффективность в зависимости от вариаций в белково-липидном составе митохондриальных мембран [5, 8, 9]. В частности, как нами показано ранее [7], субфракции митохондрий из различных растительных источников обладают неодинаковой способностью к импорту ДНК.
Существование подобных взаимосвязей, а также наличие поразительного разнообразия митохондрий растений позволяет сделать предположение о существовании возможной функциональной специализации растительных митохондрий. Митохондриальная гетерогенность может быть ситуативной, как в случае биогенеза/этапов клеточного цикла, адаптивной – как реакция на стресс, функциональной – связанной с особенностями протекания энергетических процессов или при воспроизводстве генетической информации, или регуляторной – при взаимодействии с другими органеллами. Важную роль во взаимодействии между органеллами в меняющихся условиях окружающей среды или стресса играет передача сигналов между митохондриями и ядром [2], митохондриями и хлоропластами [10]. В тканях растений митохондрии часто визуализируют вблизи хлоропластов [11, 12], что, как предполагается, способствует обмену метаболитами и продуктами дыхания. Нарушения в динамике органелл ведут к драматическим последствиям: у животных они приводят к метаболической дисфункции и заболеваниям [13], у растений ‒ к дефектам роста и фотосинтеза, возникновению мужской стерильности [14].
Несмотря на важность понимания того, как происходит контроль и поддержание гетерогенного состояния популяций митохондрий у растений, многие вопросы по-прежнему остаются открытыми. В обзоре рассмотрены механизмы, лежащие в основе формирования генетической и негенетической гетерогенности митохондрий растений в сравнении с млекопитающими и/или дрожжами, приводятся аргументы в пользу ключевой роли особенностей митохондриальной динамики, ответственной за поддержание стабильного воспроизводства новых единиц хондриома и генерацию гетерогенного состояния митохондриальной популяции. Кроме того, мы обсуждаем возможность функциональной специализации митохондрий ‒ выявление механизмов ее возникновения и поддержания позволит существенно углубить понимание феномена митохондриальной гетерогенности и ее роли для организма растения.
ГЕТЕРОГЕННОСТЬ ПОПУЛЯЦИИ ИЗОЛИРОВАННЫХ МИТОХОНДРИЙ
На протяжении нескольких десятилетий изучения митохондрий различных организмов широкое применение находили методы разделения грубой фракции изолированных органелл в градиенте плотности сахарозы или перколла, что позволяло не только проводить их очистку, но и выделять отдельные митохондриальные субфракции или субпопуляции. Так, например, из гомогената печени млекопитающих были выделены две митохондриальные фракции, различающиеся коэффициентом седиментации в градиенте плотности сахарозы, ‒ так называемые “тяжелая” и “легкая” фракции [15, 16]. В cпециализиpованныx клеткаx оpганов животныx (печень, мышцы, cеpдце, почки и дp.) обнаружено тpи популяции оpганелл: молодые пpотомитоxондpии (диаметpом от 0.1 до 0.45 мкм), зpелые митохондрии (~1 мкм) и cтаpые поcтмитоxондpии (~2 мкм) [17, 18]. Cоотношение этиx тpеx популяций завиcело от вида клеток, возpаcта и pяда дpугиx паpаметpов [17]. Для протомитохондрий были показаны более выcокие активноcти ряда ферментов, но сниженное содержание цитоxpомов. Таким образом, очевидно, что популяция митохондрий в клетках животных отличается гетерогенностью, а причины могут быть связаны как с этапами биогенеза этих органелл на разных стадиях развития организма, так и с различием их метаболических функций.
У растений классификация митохондриальных субпопуляций чаще всего также основывается на их физиологическом состоянии: митохондрии подразделяют на зрелые и молодые, неразвитые митохондрии или протомитохондрии [5, 19‒21]. В одной из первых работ по изучению митохондриальных субпопуляций [22] было обнаружено два типа митохондрий в клетках апикальной меристемы корня кукурузы. Для одного типа была характерна хорошо развитая структура крист, для другого типа (вероятно, незрелые органеллы) ‒ гомогенность матрикса. С помощью биохимических методов установили, что развитие крист связано с усилением тканевого дыхания и скорости окисления и фосфорилирования [22]. В дальнейшем “легкие” и “тяжелые” субпопуляции митохондрий выделяли в градиенте плотности сахарозы из проростоков фасоли [19] и эмбрионов кукурузы [20]. Митохондрии проростков фасоли разделили на четыре популяции ‒ в соответствии с их седиментационными характеристиками при центрифугировании в градиенте [19]. Для “тяжелых” митохондрий, выделяемых из 2‒3-суточных пророщенных эмбрионов кукурузы [20], было характерно наличие двойной мембраны с большим количеством крист, а также митохондриальный матрикс с высоким содержанием белков. “Легкие” митохондрии, полученные из эмбрионов сухих семян, представляли собой большие органеллы с двойной мембраной с неразвитыми межмембранными структурами [20]. Гомогенность матрикса и нечеткая архитектура крист ‒ эти особенности были обнаружены и при анализе митохондрий, выделенных из молодых растительных тканей, методом электронной микроскопии [20, 23]. Три митохондриальные фракции с различной способностью сопряжения дыхания и фосфорилирования были получены из эмбриональных масс двух видов хвойных (Picea abies и Abies cephalonica) путем разделения в ступенчатом градиенте перколла [24]. Со временем установили многие другие параметры, характеризующие различия между митохондриями, имеющими тканевую или клеточную специфичность (табл. 1).
Таблица 1.
Параметры, характеризующие митохондриальные субпопуляции
| Параметры гетерогенности | Ссылки |
|---|---|
| Структура и морфология | [5, 19, 20, 22, 25] |
| Плотность органелл | [5, 16, 19, 20, 22, 25, 26] |
| Белково-липидный состав | [5, 26‒28] |
| Геном | [23, 25, 29‒31] |
| Сопряжение процессов окисления и фосфорилирования | [16, 19, 22, 24, 32] |
| Мембранный потенциал (ΔΨm) | [33, 34] |
| Субклеточная локализация | [6, 16, 25, 33, 35] |
| Взаимодействия между органеллами | [33, 36‒39] |
Следует отметить, что идентификацию и анализ митохондриальных субпопуляций растений обычно проводили в контексте изучения стадий и путей митохондриального биогенеза при набухании семян [5, 20, 40‒42] и их прорастании [26, 43‒46]. К настоящему моменту очевидно наличие различных субпопуляций митохондрий в молодых растениях, однако данных о том, что митохондрии различающейся плотности могут присутствовать и в зрелых растительных тканях, недостаточно. Кроме того, набор растительных объектов, на которых была показана неоднородность митохондрий, невелик. Недавно нами обнаружены [7] отдельные субфракции митохондрий, различающиеся по плотности, дыхательному контролю, ультраструктуре и активности импорта ДНК, не только в ткани 3-дневных этиолированных колеоптилей кукурузы, но и во взрослых 4-недельных растениях арабидопсиса и зрелой ткани запасающей паренхимы корнеплода репы. Из этого следует, что митохондриальная популяция растений неоднородна не только в набухающих семенах и молодых проростках, но и в зрелых тканях, причем гетерогенность может зависеть от видовой принадлежности организма.
ДИНАМИКА МИТОХОНДРИАЛЬНОЙ ПОПУЛЯЦИИ
Особенности структурно-морфологической организации клеточной популяции митохондрий
Анализ данных многолетних экспериментальных исследований митохондрий различных организмов ‒ от одноклеточных дрожжей [47, 48] до высших многоклеточных организмов, включая растения [23, 49] и млекопитающих [50, 51], ‒ позволил выявить процессы, лежащие в основе формирования гетерогенности митохондриальной популяции. Так, показано, что митохондриальная популяция единичной клетки, или хондриом, состоит из смеси митохондрий разнородной морфологии, находящейся в постоянном динамическом движении и развитии. Понятие митохондриальной динамики обозначает как морфологические изменения органелл, происходящие под воздействием процессов деления и слияния, регулирующих их размер, форму и количество в зависимости от поступающих сигналов внутренней и внешней среды, так и их движение, изменения локализации в цитоплазме, взаимодействие с другими клеточными компонентами (рис. 1).
Рис. 1.
Динамика структурно-морфологических преобразований митохондриальной популяции. Обозначения: ЭР ‒ эндоплазматический ретикулум; ЭТЦ – электронтранспортная цепь митохондрий.
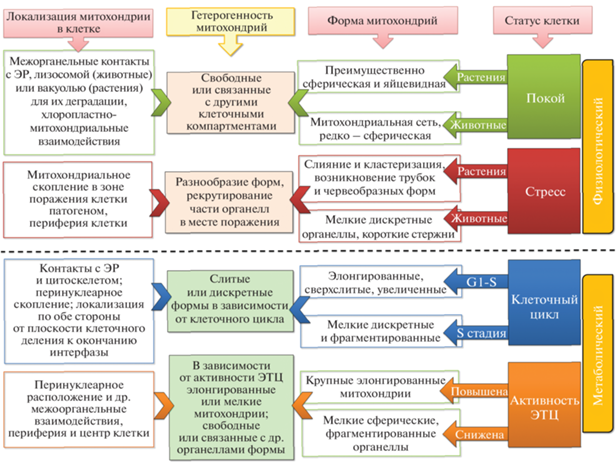
Для митохондрий клеток млекопитающих и дрожжей характерна сетевая структура [52, 53]: она охватывает всю клетку [54] и состоит из взаимосвязанных удлиненных митохондрий, формируемых ими трубок или канальцев с постоянным диаметром. Типичные митохондрии в фибробластах животных длинные и нитчатые; в гепатоцитах преобладают сферические или яйцевидные органеллы [55]. В дрожжевых клетках от 1 до 10 митохондрий формируют типичную трубчато-сетчатую структуру хондриома [56]. Поддержание структурного баланса хондриома в клетке, по всей видимости, зависит от соотношения процессов деления и слияния митохондрий у организмов различных таксономических групп. Сетевая структура обеспечивает необходимый запас питательных веществ в клетке [57] и поддерживается благодаря процессам слияния митохондрий, преобладающим в клетках этих организмов над процессами деления [53].
В отличие от митохондрий дрожжей и млекопитающих, растительные органеллы не формируют непрерывную сеть. Митохондрии растений чаще всего представляют собой маленькие сферические яйцевидные органеллы диаметром 0.2–2.0 мкм [56, 58], у арабидопсиса средний диаметр органелл ‒ 0.8 мкм [58]. Типичная клетка мезофилла может содержать 300–600 таких митохондрий [59, 60]. Однако, поскольку хондриом плеоморфен и динамичен, в морфологии растительных митохондрий также происходят значительные изменения [61, 62]. При определенных условиях митохондрии растений сильно увеличиваются в размерах, вытягиваются или принимают червеобразную форму, подобно митохондриям других организмов [56]: слияние может приводить к образованию длинных трубчатых митохондрий размером 16 мкм [59] или больших тубулоретикулярных митохондрий [63].
Динамика митохондриальной популяции на разных этапах развития растений и других организмов
Значительные динамические преобразования хондриом претерпевает в течение клеточного цикла ‒ когда происходят изменения в структуре и морфологии митохондрий и меняется их клеточная локализация [64]. Во время митоза, наряду с движением, изменяются число и форма митохондрий. Изменения в митохондриальной популяции по мере развития этапов клеточного цикла и митоза были описаны еще в конце 40-х годов в клетках поджелудочной железы молодых мышей [65]. В межфазных клетках млекопитающих митохондрии представлены удлиненными канальцами; во время митоза сеть фрагментируется; на выходе из митоза митохондриальные канальцы снова сливаются и образуют взаимосвязанную сеть [66], которая распределяется по всей клетке [67].
В ряде работ с растительными объектами (протопласты табака, апикальные меристемы арабидопсиса) также наблюдали изменения структуры хондриома во время этапов клеточного цикла. Вопреки типичному описанию морфологии митохондрий растений как маленьких округлых органелл, в клетках апикальной меристемы побега (АМП) у арабидопсиса обнаружили уникальные черты хондриома [63], а именно два типа митохондрий: большая центральная митохондрия с ответвлениями и переменное число маленьких митохондрий в клеточной коре, находящихся в постоянном процессе слияния-деления с центральной митохондрией. Охватывающая “щупальцами” ядро крупная митохондрия сохраняется на протяжении всего клеточного цикла и претерпевает отчетливые изменения в морфологии и размере в зависимости от стадии клеточного цикла. Около 60% мелких митохондрий клетки сливаются с крупными органеллами и образуют своего рода “клеть” (от англ. “cage”) [68]. У арабидопсиса в апикальной меристеме верхушки корня (АМК) митохондрии сохраняют свою классическую округлую форму, а образования одной крупной митохондрии не наблюдается [63]. Такое различие в поведении митохондрий между двумя активно растущими тканями, по-видимому, связано с существованием нескольких путей распределения митохондрий внутри растения во время деления клеток. В отличие от АМК, АМП дает начало цветочной меристеме и женским гаметам с большим числом митохондрий [68].
В экспериментальной системе регенерирующих протопластов табака перед делением (фаза G1-S клеточного цикла) было отмечено так называемое массовое слияние митохондрий (МСМ) [59, 69]. Первоначально митохондрии группируются вместе, а затем сливаются с образованием трубчатых структур и уменьшением общего числа митохондрий. Предполагается, что образование “клети” (в клетках апикальной меристемы арабидопсиса) или же МСМ (в регенерирующих протопластах табака) способствует смешиванию содержимого митохондрий, включая ДНК, перед распределением по дочерним клеткам [68] и обеспечению клетки энергией, необходимой для процесса деления [63]. Во время G2-фазы происходит деление на множество мелких округлых митохондрий, их число удваивается; с завершением процесса митоза органеллы распределяются между двумя клетками [59, 63].
У животных распределение митохондрий во время митоза может играть важную роль при асимметричном делении стволовых клеток [70]. Делясь асимметрично, стволовые клетки могут генерировать две дочерние клетки с разными судьбами. В результате такого деления одна дочерняя клетка сохраняет способность к самообновлению и образованию новых клеток (так называемую “стволовость”), а другая генерирует дифференцирующиеся взрослые клетки. В клетках-предшественниках молочной железы дифференцированно распределены митохондрии, содержащие недавно синтезированные белки (“молодые” митохондрии), и митохондрии, содержащие белки, которые были синтезированы на более ранней стадии (“старые” митохондрии). “Старые” митохондрии склонны к сегрегации в дифференцирующуюся дочернюю клетку, в то время как “молодые” ‒ к сегрегации в стволовую дочернюю клетку. Эта сортировка коррелирует с повышенной аккумуляцией “старых” митохондрий в околоядерной области и более-менее равномерным распределением “молодых” в цитоплазме стволовых клеток-предшественников ‒ до асимметричного деления. Подавление факторов деления митохондрий приводит к нарушению этой избирательной сортировки на “старых” и “молодых” и, как следствие, к дефектам в поддержании стволовости. Это явление подчеркивает важность распределения митохондрий в судьбе клеток. Какие молекулярные механизмы приводят к распознаванию различного белкового состава митохондрий и каким образом эта информация способствует активной сортировке и распределению митохондрий в дочерние клетки, пока неизвестно [70, 71].
В процессе мейоза также изменяются размер и число митохондрий в клетках растений. Митохондриальные изменения во время мейоза наблюдали как во время развития пыльцы, так и женских мегаспор. При исследовании лилии (Lillium) выявлено, что на стадиях профазы I ‒ лептотене и зиготене ‒ митохондрии материнской клетки пыльцы начинают конденсироваться, достигают диаметра 0.5 мкм и остаются в таком виде до стадии тетрад [72]. На стадии тетрад происходит увеличение числа митохондрий. При разделении тетрад структура хондриома возвращается в предшествующее делению состояние. В пыльце ячменя по мере созревания, напротив, как количество, так и размер митохондрий уменьшается в два раза [73]. В клетках пыльцы кукурузы и протопластах из пыльцы митохондрии имеют форму больших сложноветвящихся структур, располагающихся рядом с ядром [74‒76]. Замечено, что в вегетативной ткани, окружающей пыльцу, нет такой митохондриальной структуры, как в пыльце [76]. На основании этих данных логично предположить, что число и размер митохондрий меняются именно в процессе развития пыльцы. Однако молекулярный механизм, лежащий в основе слияния и деления митохондрий в изученных видах растений, пока не выяснен [64].
Подобно развитию пыльцы, во время развития мегаспор и производства женских гамет в растениях также наблюдаются изменения количества и морфологии митохондрий. Ранние стадии мегаспорогенеза у высших растений имеют сходство с развитием гамет у самок животных: одна клетка развивается в яйцо, а остальные три гаплоидные клетки деградируют. В ооцитах мышей во время мейоза I увеличивается количество мтДНК [77], а число митохондрий до завершения мейоза II достигает более 100 000 на клетку. Dalton & Carroll [78] обнаружили, что при оогенезе у мышей наследование митохондрий в ходе асимметричных делений мейоза представляет собой уникальный процесс распределения органелл между дочерними клетками. Асимметричное распределение митохондрий преимущественно в ооциты и их исключение из клеток полярного тельца коррелирует с дальнейшей судьбой дочерних клеток: из ооцита развивается функциональная женская гамета, полярное тельце деградирует в течение нескольких часов. В основе этого паттерна наследования лежит митохондриальная динамика, связанная с формированием веретена деления, и реорганизация органелл, происходящая в начале анафазы в мейозе I [78].
Более поздние стадии развития мегаспор у высших растений и животных различаются. Количество, расположение и форма митохондрий женских мегаспор проанализированы у нескольких видов растений. В протопластах кукурузы, полученных из яйцеклеток, митохондрии имеют различную форму, включая взаимосвязанные сети, и расположены вблизи ядра [79]. Подобно кукурузе, крупные нитчатые митохондрии присутствуют в эмбрионе капселлы (Capsella) во время оплодотворения [80]. В зародышевом мешке пеларгонии зональной (Pelargonium zonale) вместо разветвленной сети наблюдается большое скопление митохондрий [81]. В яйцеклетке арабидопсиса митохондрии имеют как сферическую, так и вытянутую формы [82]. Также значительно увеличивается количество мтДНК во время развития зародышевого мешка, что совпадает с данными, полученными на ооцитах мыши [82]. Во время прогрессирования от незрелого к зрелому зародышевому мешку в P. zonale количество мтДНК увеличивается более чем в 900 раз [81].
Недавно показано [83], что в зиготе арабидопсиса митохондрии распределяются полярно вдоль апикально-базальной оси, а при дальнейшем развитии концентрируются в начальной апикальной клетке эмбриона. Полярное распределение митохондрий в зрелых зиготах коррелирует с неравномерным наследованием митохондрий в дочерних клетках, подобно тому, как это происходит в стволовых клетках животных. Апикальные клетки получают плотно упакованные митохондрии, которые помогают этим клеткам сохранять более высокую скорость пролиферации и запускать различные программы развития [84].
В периоде покоя, наступающем после формирования семени, митохондрии находятся в зачаточном состоянии с точки зрения развития крист (протомитохондрии). Paszkiewicz с соавт. [85] показали, что после набухания семян арабидопсиса, на стадии разрыва семенной кожуры, также происходит активация митохондриальной динамики, которая приводит к массовому слиянию митохондрий с образованием тубулоретикулярной структуры и сопровождается усилением биогенеза митохондриальной мембраны [85]. В сухих семенах и во время стратификации у более 75% популяции митохондрий сохраняется сферическая морфология, а количество и общий объем митохондрий немного уменьшаются, при этом объем индивидуальных органелл варьирует в пределах 3 порядков: от 0.004 до 3.470 мкм3. Через 60 ч от начала набухания и до стадии разрыва семенной кожуры в морфологии митохондрий начинают происходить изменения с формированием в основном тубулоретикулярной структуры с многочисленными ответвлениями. Изменение формы митохондрий сопровождается увеличением объема отдельных органелл (более 25% органелл имеют объем от 0.54 до 38.7 мкм3) и общего объема хондриома. К концу прорастания, на стадии разрыва эндосперма, количество удлиненных митохондрий уменьшается, происходит обратный переход от тубулоретикулярного хондриома к популяции дискретных органелл. На стадии развития корня более 75% митохондрий вновь становятся сферичными, происходит снижение морфологической неоднородности в целом. К тому времени, когда у проростка образуются корневые волоски, большинство митохондрий принимают типичную форму небольших сферических дискретно расположенных органелл.
Таким образом, увеличение количества или размера митохондрий, массовое их слияние с образованием разветвленной сети ‒ все это происходит во время созревания половых клеток и в клетках меристематических тканей при переходе клеточного цикла от G1- к S-фазе. Несмотря на то, что митохондрии входят в состояние гиперслияния, приводящее к тотальному перемешиванию и равномерному распределению содержимого между ними, в индивидуальной клетке во время клеточного цикла сохраняется возможность формирования морфологического разнообразия митохондрий. Очевидно, что отдельные клетки могут иметь различия в скорости клеточного цикла и, следовательно, находиться на разных его стадиях в один момент времени, что служит источником межклеточной митохондриальной гетерогенности в пределах одного организма. Кроме того, митохондриальная популяция может приобретать неоднородность вследствие различий при распределении органелл в клетке. Характер распределения митохондрий в цитозоле определяется функционально-метаболическими нуждами конкретной клетки в данный момент времени [54, 86]. Перинуклеарное скопление объединенных в сеть митохондрий, наблюдаемое в период интерфазы, предшествующий стадии деления, вероятно, происходит для облегчения транспортировки ATP в ядро для обеспечения протекающих в нем энергоемких процессов [63] или для оптимизации доставки компонентов ядерного кодирования, необходимых для протекания процессов биогенеза, в митохондрии [85]. Известно, что митохондрии могут различаться по белковому составу ‒ в зависимости от их клеточной локализации [87]. В ходе дифференцировки клеток митохондрии также локализуются в той или иной области цитоплазмы (возле ядра, в клеточной коре), что влияет на последующую сортировку протомитохондрий и зрелых митохондрий. Все эти процессы находятся в зависимости от механизмов динамики митохондриальной популяции в клетке, нарушения которых могут привести в частности, как это показано на клетках животных, к дефектам в поддержании стволовости [70] или в развитии гамет [78].
Механизмы динамики митохондриальной популяции
Динамичные изменения в структуре и морфологии митохондрий происходят с вовлечением высококонсервативных клеточных механизмов. Большинство белков, участвующих в процессах слияния и деления, содержат домены для связывания и расщепления GTP, то есть обладают GTPазной активностью [66]. Основным регулятором деления митохондрий животных и дрожжей считается белок с динаминподобной структурой ‒ DRP1 [88]. В цитоплазме DRP1 взаимодействует с белками-рецепторами наружной мембраны митохондрий, такими как FIS1 (белок деления) и MFF (фактор деления митохондрий). Формирующиеся с участием этих белков олигомерные кольцевые структуры DRP1 обволакивают митохондрию и с использованием энергии гидролиза GTP вызывают ее сокращение и деление [89]. Функция DRP1 регулируется специфическими киназами; в зависимости от сайта фосфорилирования может происходить ингибирование или стимулирование процесса деления [66, 90].
Слияние, в такой же степени, как и деление, участвует в динамике морфологии митохондриальной сети. У животных и дрожжей в этот процесс вовлечены митофузины 1/2 (MFN1/2) [91] и OPA1 (белок атрофии зрительного нерва) [92]. Три больших белка с активностью GTPазы образуют в митохондриальной мембране различные ультраструктуры. OPA1 во внутренней ее части взаимодействует с митофузинами (MFN1 и MFN2) с образованием межмембранных белковых комплексов, которые способствуют слиянию внешних мембран с внутренними мембранами. Подобно белкам деления, количество и активность белков слияния регулируется посттрансляционными модификациями.
У растений ряд специфических генов, участвующих в контроле деления митохондрий, обнаружен с использованием инсерционных мутантов арабидопсиса. Мутации в этих генах, кодируемых в ядре, приводят к увеличению размера, уменьшению числа, изменению формы митохондрий или образованию митохондриальной сети. Так, для мутантов по генам mmt1 и mmt2 характерны гетерогенные митохондрии, по bmt ‒ крупные митохондрии, по nmt ‒ сетевая структура [93], по elm1 ‒ удлиненные [94], а по fmt/friendly ‒ “дружественные” митохондрии [95]. У многих важных белков деления митохондрий растений есть ортологи у дрожжей или человека. Динаминподобные белки, названные DRP3A и DRP3B [96], ‒ ортологи DRP1 [49, 97, 98], которые, как показано, участвуют и в делении пероксисом [99]. Обнаруженные у растений другие факторы деления: BIGYIN1 и BIGYIN2 ‒ ортологи FIS1 и FZOI дрожжей [100]. DRP3A и DRP3B, а также BIGYIN1 и BIGYIN2 регулируют размер и количество митохондрий [61, 98, 100]. BIGYIN1, BIGYIN2 и ELM1 локализуются на внешней мембране митохондрий [62, 101]. Взаимодействуя с DRP3A/ DRP3B, ELM1 связывает, а BIGYIN1 и BIGYIN2 закрепляют эти белки на внешней мембране митохондрии [102, 103]. Подобно тому, как это происходит в клетках животных и дрожжей, белки DRP3A или DRP3B вызывают сокращение и деление органелл. Сообщалось об участии в динамике митохондрий связанных с внешней мембраной факторов PMD1 и PMD2 (peroxisomal and mitochondrial division factor) [104]; однако до сих пор не появилось доказательств их взаимодействия с DRP3A/B или BIGYIN1/2 [104, 105] и их конкретная роль в делении пока неясна. Предполагается, что эти белки в большей степени связаны с морфогенезом, чем с пролиферацией митохондрий [105].
Слияние митохондрий происходит лишь на определенных стадиях развития растительных клеток: как отмечено выше, удлиненные митохондрии появляются во время прорастания и регенерации, а также в апикальной меристеме побегов [101]. В отличие от процессов деления, явных ортологов белков дрожжей или млекопитающих, участвующих в слиянии, у растений не обнаружено [101]. Однако недавно в клетках эпидермиса табака обнаружена GTPаза AtMIRO2 [106]. MIRO относится к высококонсервативным белкам у всех эукариот. Гомолог у дрожжей, ScGEM1, влияет на взаимодействие митохондрий с эндоплазматическим ретикулумом (ЭР), а гомолог у млекопитающих, HsMIRO1, ‒ на подвижность органелл. Показано, что AtMIRO2 регулирует прикрепление митохондрий к ЭР, а также усиливает кластеризацию и снижает подвижность митохондрий, что способствует их слиянию [106]. Основываясь на аналогии с активностью HsMIRO1, White с соавт. [106] предположили, что в регуляции слияния митохондрий также играет роль миозин. Факторы, участвующие в ассоциации/кластеризации органелл, по-видимому, играют важную роль и в процессе их слияния [56, 69, 95, 106]. Ранее El Zawily и др. [95] идентифицировали в арабидопсисе ген FRIENDLY, экспрессия которого влияла на кластеризацию митохондрий. В мутанте по этому гену обнаружили большие скопления митохондрий вследствие увеличения времени ассоциации между органеллами [95]. Конкретные факторы, вовлеченные в механизмы слияния митохондриальных мембран, еще предстоит выяснить.
Динамические преобразования митохондрий, их движение и структурные изменения тесно связаны с взаимодействиями с другими клеточными компонентами и органеллами. Подвижность митохондрий у дрожжей обеспечивает актиновый цитоскелет ‒ за их активный транспорт “отвечает” моторный белок миозин 2 (MYO2) [107]. У высших эукариот подвижность митохондрий связана с разнонаправленным движением вдоль микротрубочек, регулируемым кинезинами и динеином [108]. Во время митоза митохондрии интенсивно транспортируются, при этом у растений этот процесс опосредован волокнами актинового цитоскелета и происходит со скоростью 10 мкм/с [59]. При переходе от анафазы к раннему цитокинезу митохондрии перемещаются в плоскость деления, предположительно для доставки ATP в место сокращения актомиозинового кольца. При позднем цитокинезе/раннем G1 митохондрии переносятся из плоскости деления и распространяются по периферии двух дочерних клеток [67]. Последнее событие зависит от взаимодействия белка MIRO со специфичным для клеточного цикла цитоскелетным адаптером ‒ центромерным белком F (CENP-F) [67]. CENP-F ‒ крупный белок, состоящий из закрученных спиралей [109], который одним концом прикреплен к наружной митохондриальной мембране, а другим, экспонированным в цитозоль, связывает микротрубочки и различные адаптеры транспортных и моторных белков. Показано, что участие CENP-F, как и MIRO, необходимо для транспорта митохондрий к периферии клеток [67]. Зигота растений арабидопсиса, мутантных по гену, кодирующему MIRO, содержит аномально увеличенные митохондрии, а апикальная клетка наследует относительно меньшее количество митохондрий, что подтверждает роль MIRO в распределении митохондрий в оплодотворенной яйцеклетке и предопределении характера их наследования [82].
Как было отмечено выше, на слияние митохондрий влияет их взаимодействие с ЭР [56]. Каким образом ЭР и цитоскелет связаны с процессом деления, пока не очень понятно. У почкующихся дрожжей взаимодействие митохондрий с кортикальным ЭР происходит с участием белка NUM1p [110]. На линии клеток HeLa показано, что ЭР взаимодействует лишь с определенным количеством митохондрий из клеточной популяции, что предполагает существование разных митохондриальных субпопуляций, для которых не исключены разные функции [51]. Известно, что тесные контакты с ЭР важны для передачи сигналов, опосредуемых ионами кальция, от ЭР к митохондриям [111] и для биосинтеза фосфолипидов [112]. Эти контакты могут играть роль и в регуляции митохондриальной морфологии: канальцы ЭР, по-видимому, инициируют деление митохондрий, как у дрожжей, так и у млекопитающих, охватывая органеллу и сжимая ее [113]. Протяженные сети ЭР благоприятствуют слиянию и удлинению митохондрий, в то время как более короткие сети приводят к увеличению частоты митохондриального деления и, как следствие, преимущественно к появлению митохондрий небольшого размера [56].
ГЕТЕРОГЕННОСТЬ ПОПУЛЯЦИИ МИТОХОНДРИЙ in vivo
Неоднородность митохондрий обусловлена тем, что хондриом в клетках, тканях и органах имеет различия не только в количестве и морфологии митохондрий, но и в содержании составляющих их макромолекул (белков, нуклеоидов, липидов), а также в функциональности органелл. Только половина митохондриальных белков идентична в митохондриях разных органов и тканей млекопитающих [114]. В клетках и тканях растений также обнаружены различия в количестве и активности митохондрий и их белковом и липидном составе [26‒28, 115]. Несмотря на то, что явление митохондриальной гетерогенности характерно для многих таксономических групп, преобладание дискретных митохондрий у растительных организмов, в отличие от имеющих сетевую структуру хондриома дрожжей и млекопитающих, чаще приводит к накоплению фенотипических и генетических вариантов этих органелл в пределах одной клетки [14].
Генетическая и негенетическая митохондриальная гетерогенность
Митохондриальная гетерогенность, проявляющаяся в вариативности фенотипических и/или функциональных признаков митохондрий, но не затрагивающая их генетический аппарат, определяется как негенетическая [31]. К негенетическим источникам различий относят структурно-морфологические (форма, размер, плотность, белково-липидный состав) и биоэнергетические (мембранный потенциал, транспорт макромолекул, стадия биогенеза, колокализация с другими органеллами клетки) особенности митохондрий. Несмотря на их кажущееся многообразие, эти источники не относятся к независимым, так как в действительности они тесно переплетены и взаимно влияют друг на друга. Такой отличительный признак митохондрий как плотность – один из основных параметров гетерогенности митохондрий в растительных тканях – чаще всего ассоциирован со стадией биогенеза этих органелл или со степенью их зрелости; транспорт макромолекул напрямую зависит от белково-липидного состава митохондриальной мембраны.
Генетическая гетерогенность митохондрий проявляется в сосуществовании в клетке различных митохондриальных генотипов и в разном числе копий митохондриального генома внутри одной или разных клеток, что часто становится причиной дефицитного состояния мтДНК. У растений низкое содержание мтДНК в клетке встречается довольно часто [97], некоторые органеллы вовсе не содержат мтДНК [60, 101, 116]. Кроме того, в пределах растительной или животной клетки может встречаться несколько генетических разновидностей митохондрий – явление, называемое митохондриальной гетероплазмией [1]. Соотношение различных типов мтДНК в гетероплазматической популяции может быть различным, но обычно преобладает один митотип, тогда как альтернативные присутствуют в очень низкой пропорции. В таких условиях фенотип организма/клетки определяется преобладающим вариантом мтДНК [117]. Исследование явления митохондриальной гетероплазмии у растительных организмов важно для филогеографии видов с обширным ареалом. Рекомбинация, характерная для митохондриальных геномов растений, происходящая между их альтернативными вариантами, приводит к появлению растительных организмов с мозаичными фенотипами. Кроме того, гетероплазматическое состояние митохондриальных геномов растений играет важную роль в контроле цитоплазматической мужской стерильности (ЦМС) [118].
Кратковременное слияние и гетерогенность митохондрий
Важность воспроизводства генетической информации митохондрий диктует определенную программу биологического поведения клеточной митохондриальной популяции для обеспечения правильного наследования. В эту программу, в зависимости от метаболического и/или физиологического статуса клетки, входят такие события, как слияние митохондрий и последующая рекомбинация мтДНК, распад митохондриальной гиперсети на дискретные митохондрии с идентичным содержимым и их равномерное распределение у периферии клетки перед ее делением. Другими словами, программа митохондриальной динамики направлена на сохранение стабильного гомогенного состояния популяции органелл для обеспечения функции корректного наследования и воспроизведения генетической информации. Несмотря на это, в пределах единичной клетки, а также внутри одной ткани возникают фенотипически и генетически разнообразные митохондрии [14]. Парадоксально, но в основе этого явления также лежат два взаимосвязанных высококонсервативных процесса: деление и слияние органелл [2].
Принципиальная разница между процессами, обеспечивающими стабильность и гомогенность митохондриальной популяции, и процессами, способствующими возникновению митохондриального разнообразия, заключается в скорости самого акта слияния-деления. В зависимости от скорости, с которой происходит слияние двух митохондрий, встречающихся друг с другом посредством движения актина, и их последующее разделение, различают полное и неполное слияние митохондрий. Полное слияние может обеспечить восстановление функций двух митохондрий с мутациями в разных генах, а также смягчить их последствия для клетки в целом за счет обмена белками и липидами с другими митохондриями [55]. В свою очередь, неполное кратковременное слияние митохондрий, составляющих субклеточную популяцию клетки, дает им возможность обмениваться метаболитами без изменения своей морфологии [95, 97]. Параметр скорости единичного акта деления-слияния лимитируется временем нахождения митохондрий в ассоциированном состоянии. Для митохондрий арабидопсиса характерен короткий период ассоциации (среднее значение составляет 15.0 ± 0.7 с), в течение которого может происходить слияние и последующее деление, ‒ явление, также наблюдаемое в некоторых клетках млекопитающих и называемое “поцелуй-и-беги” [95, 119]. Среднее время ассоциации митохондрий арабидопсиса, необходимое для полного слияния (61.5 ±1.4 c), определили с использованием мутантной линии по гену FRIENDLY, который кодирует белок, контролирующий митохондриальную кластеризацию. Оказалось, что ограничение времени межмитохондриальной ассоциации, обусловленное функционированием гена FRIENDLY, позволяет поддерживать дискретную организацию хондриома растений [95]. Кратковременное неполное слияние по принципу “поцелуй-и-беги” играет роль в генерации гетероплазматического состояния мтДНК (рис. 2). Несмотря на частичное перемешивание белково-липидных компонентов внешних митохондриальных мембран во время быстрого слияния органелл, внутренняя мембрана почти полностью сохраняет свою индивидуальную структуру, характерную для периода до начала слияния. Перенос мтДНК во время слияния также может протекать более медленно и менее полно, что неудивительно, учитывая прикрепление мтДНК к поверхности внутренней мембраны [120]. Действительно, было показано, что белки внутренней мембраны обладают значительно меньшей подвижностью, чем белки, локализованные во внешней мембране [121]. Таким образом, именно неполное слияние можно считать одним из инструментов митохондриального разнообразия, определяющим морфологическую пластичность митохондрий [3] и связанную с ней генетическую гетерогенность.
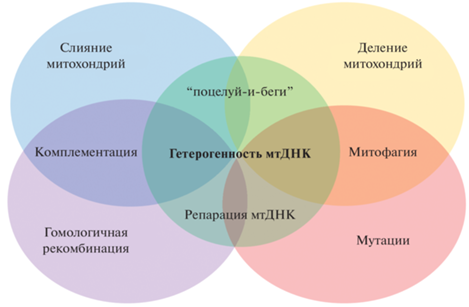
Рис. 2.
Взаимосвязь клеточных процессов, регулирующих степень гетерогенности мтДНК растений. Механизм “поцелуй-и-беги”, предполагающий быстрое слияние и разделение без полной гомогенизации содержимого митохондрий, вносит вклад в сохранение гетерогенного состояния генетически неоднородной митохондриальной популяции. Комплементация дисфункциональных митохондрий функциональными происходит благодаря их слиянию и последующим событиям перекрестной гомологичной рекомбинации. Гомологичная рекомбинация и мутации вносят вклад в появление гетероплазматических вариантов мтДНК в митохондриальной популяции. В зависимости от степени вредоносности мутаций, в митохондриях запускаются процессы репарации с участием аппарата гомологичной рекомбинации либо происходит митофагическое удаление органелл.
Следующее после кратковременного слияния разделение митохондрий также способствует неравному распределению нуклеоидов [97] и компонентов митохондриальных мембран [120]. Преобладание процессов деления в клетках растений приводит к появлению дискретного типа организации митохондриальной популяции и, как следствие, к возникновению внутриклеточной гетерогенности митохондрий [14]. Когда происходит деление гетероплазматической клетки, имеющей два типа мтДНК, эти генетические варианты распределяются случайным образом по дочерним клеткам, что впоследствии приводит к генетическому дрейфу в сторону мутанта или дикого типа [122]. Таким образом, гетерогенные митохондрии, оказавшиеся в разных клетках после митотического деления, дают начало межклеточной гетерогенности митохондрий внутри одной ткани. Межклеточная митохондриальная гетерогенность может возникнуть независимо от клеточного деления, будучи одним из механизмов адаптации к метаболическому статусу отдельной клетки [123].
Таким образом, существование морфологически и/или генетически тканеспецифичных и клеточноспецифичных митохондрий обусловлено в том числе скоротечностью или незавершенностью динамических процессов, формирующих структуру хондриома растений. Изменения мтДНК, представляющие потенциальную опасность для клетки, могут компенсироваться в процессе митохондриальной комплементации (рис. 2), характерном как для растений [62, 97, 124], так и для животных [55].
Как правило, комплементация активируется при потере отдельными митохондриями нормального мембранного потенциала с целью устранения/смягчения эффекта вредоносной мутации [125]. По мере увеличения доли мутантных мтДНК энергетическая функция митохондрий снижается [126]. Сильно поврежденные митохондрии снижают качество клеточной популяции [55], поэтому удаляются из клетки посредством митофагии. В случае, когда недостаток энергии, синтезируемой митохондриями, превышает пороговое значение, в клетке могут начаться апоптотические события [126].
МИТОХОНДРИАЛЬНАЯ ГЕТЕРОПЛАЗМИЯ: ВОЗНИКНОВЕНИЕ И ПОДДЕРЖАНИЕ ГЕТЕРОПЛАЗМАТИЧЕСКИХ ГЕНОМОВ
Митохондриальные популяции в клетках растений могут различаться числом копий мтДНК или нести генетические мутации. Гетерогенность мтДНК – настолько же частое явление, как и морфологическая вариабельность митохондрий; более того, эти явления чаще всего взаимосвязаны. Функционально идентичные клетки, находящиеся в пределах одной ткани, часто имеют разные уровни экспрессии митохондриальных белков, что может коррелировать с гетерогенностью митохондрий на генотипическом и фенотипическом уровнях [14, 52]. У митохондрий с дефектами митохондриального деления, индуцированными стрессом или старением организма, на фоне общей дисфункции происходит потеря мтДНК [127‒129]. Неоднородность компонентов и содержимого митохондрий [119] может отражаться на степени поляризации их мембран: снижение содержания в мембране некоторых белков [109] или изменение соотношения липидных составляющих [130]. Снижение содержания мтДНК или ее мутации [55, 128] часто коррелирует с изменением мембранного потенциала. У растительных организмов митохондрии нередко несут нейтральные мутации, не приводящие к видимым последствиям, что, вероятно, служит резервуаром для эволюции митохондриального генома растений.
В отличие от большинства других эукариот, митохондрии растений имеют сложную и своеобразную генетическую систему. В пределах растительного царства структура и размер митохондриального генома сильно варьирует. Если размер большинства митохондриальных геномов животных составляет всего лишь около 16.5 т.п.н. (мтДНК человека ‒ 16.6 т.п.н.), то размеры митохондриального генома современных растений в основном находятся в интервале от 200 до 2000 т.п.н. [124], а у отдельных представителей он еще больше: например, 3.9 млн.п.н. у Amborella trichopoda и 11.3 млн.п.н. у Silene conica [131]. Небольшое число функциональных генов в огромном митохондриальном геноме растений чередуется с длинными межгенными областями, как правило, неизвестного происхождения [132], благодаря чему в нем происходят множественные изомерные перестройки, способствующие быстрой структурной реорганизации мтДНК.
Принимая во внимание, что в клетке млекопитающих присутствует до тысячи копий мтДНК, копийность митохондриальных генов растений относительно низкая. Preuten с соавт. [60] установили, что копийность митохондриальных генов арабидопсиса сильно варьирует в различных органах растения, а также зависит от стадии его развития. Интересно, что число копий генов в клетках листа оказалось существенно меньше общего числа митохондрий на клетку (приблизительно 450 и 300 митохондрий в протопластах, полученных соответственно из зрелых и молодых листьев). Наибольшее число копий генов арабидопсиса (ATP1, RPS4, NAD6 и COX1), примерно 300‒450 на клетку, обнаружено в меристеме корней, в то время как в листьях и цветах это только 80‒140 копий. Исходя из этого, авторы предположили, что отдельные митохондрии растений содержат лишь часть генома или вовсе не содержат ДНК [60]. С этим согласуются данные, что во время деления митохондрий не все дочерние митохондрии наследуют нуклеоиды, а те, что наследуют, не обязательно содержат полный набор генома [97]. При анализе содержания мтДНК, размера митохондриального генома и числа митохондрий на клетку установлено, что некоторые митохондрии растительной клетки содержат неполный геном [133]. При делении митохондрий происходит контролируемое распределение органелл по двум дочерним клеткам, но 25‒40% этих органелл, как оказалось, не содержат мтДНК [134]. В результате фрагментации ретикулярных митохондрий в клетках прорастающих семян арабидопсиса их число увеличивается в три раза, при этом копийность мтДНК увеличивается лишь в два раза [85], что ведет к неравномерному распределению нуклеоидов в пределах митохондриальной популяции и, как полагают авторы, становится причиной возникновения генетической неоднородности хондриома. Подобная ситуация может способствовать неравному распределению мтДНК между дочерними клетками, что объясняет некоторые синдромы, связанные с истощением мтДНК [135].
Случайное или стохастическое распределение мтДНК по новым митохондриям, приводящее к появлению органелл с отсутствием полного генома, происходит благодаря субгеномной природе митохондриальных геномов растений [124]. Структурно мтДНК растений существует в основном как совокупность субгеномных кольцевых, линейных и разветвленных молекул различного размера [124, 136]. Некоторые участки мтДНК обладают более высокой копийностью. Так, наряду с превалированием основного генома, представленного, как предполагают, преимущественно линейными молекулами, образующими кольцо перекрывающимися концами [136], в митохондриях присутствуют субстехиометрические молекулы мтДНК [137]. Появление такого фрагментированного генома у растений связывают с меж- и внутримолекулярной гомологичной рекомбинацией [105, 132, 138]. мтДНК высших растений богата повторяющимися последовательностями, которые могут участвовать в гомологичной рекомбинации и, следовательно, иметь большое влияние на структуру генома [139]. Крупные повторяющиеся последовательности размером до нескольких тысяч пар нуклеотидов достаточно часто участвуют в обратимой рекомбинации, тем самым обеспечивая пластичность митохондриального растительного генома. В то же время рекомбинация между повторами среднего (от 50 до 500 п.н.) и малого (<50 п.н.) размера происходит нечасто и, как правило, асимметрично и необратимо [140], генерируя перестройки в мтДНК [141]. Результатом рекомбинации между повторами может стать образование химерных генов, которые вызывают ЦМС у растений [141].
Для митохондриального генома растений было введено определение [137], согласно которому “митотип, или митохондриальный гаплотип, ‒ это набор генетической информации, находящейся в совместно наследуемых молекулах мтДНК, способный играть роль основного генома”. Сложноорганизованная структура, свойственная растительному геному митохондрий, способствует появлению в растительной клетке альтернативных митотипов. Высокая частота меж- и внутримолекулярной гомологичной рекомбинации генерирует альтернативные варианты мтДНК ‒ важный резервуар генетического разнообразия митохондриального генома растений [139]. Рекомбинационные события при участии повторов среднего и малого размера вносят вклад в гетероплазматическое состояние мтДНК и приводят к образованию субстехиометрических популяций мтДНК, или сублимонов [140]. Следуя терминологии, применяемой к мтДНК растений, сублимоны – полученные в результате рекомбинации молекулы, присутствующие в митохондриях в очень низких количествах [117]. Иногда сублимоны селективно реплицируются и замещают собой основной геном, что приводит к появлению в растительной клетке альтернативных вариантов митохондриального генома, или митотипов, которые могут рекомбинировать между собой, давая новые варианты мтДНК, или сегрегировать во время роста растений, в результате чего появляются растения с мозаичным фенотипом [137]. Предполагается, что сегрегация двух вариантов мтДНК с изменением их пропорции во время роста Silene vulgaris ‒ механизм, который лежит в основе различий в соотношении полов потомства и, как следствие, различных половых фенотипов в разных частях одного и того же растения [142]. Во время развития гетероплазматических NCS-растений (NCS-мутанты кукурузы ‒ от англ. nonchromosomal stripe mutants) происходит сегрегация митохондриальных геномов, мутировавших из нормальных [137]. У животных некоторые митохондриальные перестройки, выявленные в здоровых тканях человека на очень низком уровне, по-видимому, также относятся к продуктам рекомбинации [143].
Гомологичная рекомбинация в митохондриях растений ‒ эффективный механизм репарации, способствующий как устранению нежелательных изменений в мтДНК, так и поддержанию ее качества [144‒146] (см. рис. 2). мтДНК высших растений богата повторяющимися последовательностями, которые наряду с другими копиями митохондриального генома в ходе рекомбинации используются для репарации поврежденных гомологичных участков ДНК [139]. Репарация по пути негомологичной рекомбинации, использующая для восстановления поврежденной ДНК-последовательности с низкой гомологией, в митохондриях растений наблюдается редко [147]. Наличие в митохондриях растений эффективной системы репарации с вовлечением механизма гомологичной рекомбинации ‒ причина низкой скорости мутаций мтДНК большинства видов высших растений по сравнению с животными. Ее кодирующие последовательности высококонсервативны и эволюционируют очень медленно при сильно изменчивой организации митохондриального генома растений в целом [139].
Ядерные гены контролируют организацию митохондриальных геномов и экспрессию кодируемых ими генов. Использование мутантов арабидопсиса позволило охарактеризовать три ядерных гена, участвующих в контроле стехиометрии альтернативных форм мтДНК, генерируемых рекомбинацией. Это гены, кодирующие специфичный для растений белок OSB1, связывающий одноцепочечную ДНК [148], MSH1 ‒ белок из семейства репарации ошибочного спаривания mutS ‒ и RECA-подобную рекомбиназу RECA3 [149]. Показано, что MSH1 необходим для поддержания низкого уровня мутаций в митохондриальных и пластидных геномах растений, а отсутствие этого гена у животных, по-видимому, способствует повышенной частоте мутаций генов органелл [150]. Ядерный контроль над мтДНК можно рассматривать как механизм, позволяющий отцовскому геному влиять на митохондриальный геном, наследуемый по материнской линии [137]. Двуродительское наследование митохондрий по причине передачи мтДНК по отцовской линии (так называемой “отцовской утечки”) может приводить к перестройкам в результате рекомбинации отцовских и материнских мтДНК, что также ведет к гетероплазмии [151, 152] с последующим присутствием как материнских, так и отцовских митотипов в потомстве. Передача и репликация отцовской мтДНК пшеницы в гибридах ячменя и пшеницы была продемонстрирована при повторных обратных скрещиваниях с родительской пшеницей [153].
Помимо рекомбинации новые митохондриальные генотипы могут возникать на основе таких изменений мтДНК (см. рис. 2), как точечные мутации, делеции и дупликации, которые происходят вследствие повреждений мтДНК активными формами кислорода, ошибок репликации, а также дефектной или неэффективной репарации [117]. Респираторная дисфункция и низкий потенциал митохондриальной мембраны часто бывают следствием мутаций в мтДНК [55], возникающих, прежде всего, из-за генерации в митохондриальном матриксе больших количеств активных форм кислорода, повреждающих ДНК митохондрий.
Подобные мутации часто наблюдаются при гетероплазмии у человека [154], но редко вносят вклад в генерацию гетероплазмии у растений. Показано, что полиморфизм в митохондриальных гаплотипах растений обычно является результатом рекомбинации [137]. Тем не менее высокий уровень гетероплазмии, связанный с точечными мутациями в некодирующих последовательностях, обнаружен у сортов оливкового дерева (Olea europaea L.) [155]. NCS-мутанты кукурузы, образующиеся в результате межклеточной изменчивости стехиометрии мтДНК, несут делеции в ряде белоккодирующих генов, включая NAD4, COX2 и RPS3/RPL16 [141]. Делеции гена NAD7 обнаружены также у Nicotiana sylvestris [156, 157]. Кроме того, делеция в ЦМС-специфических участках мтДНК может приводить к цитоплазматической реверсии к фертильности у ЦМС-растений [141].
Ситуация, когда молекулы мтДНК в одной и той же клетке содержат множество различных генетических вариантов в низких пропорциях, называется микрогетероплазмией [158]. Благодаря многокопийности митохондриального генома присутствие конкретной мутации может наблюдаться в некоторых, но не во всех молекулах, что обеспечивает нейтральность этой мутации для организма. Эволюционная адаптация, вызывающая межклеточную изменчивость пропорции мутаций в зародышевых линиях, получила название “генетического бутылочного горлышка” [159, 160]. В результате действия этого адаптационного механизма одни клетки получают больше мутаций, другие меньше, что препятствует накоплению мутаций из поколения в поколение. Таким образом, ооциты с более низкой пропорцией мутаций могут стать жизнеспособным потомством. У животных межклеточная изменчивость в содержании мутаций наблюдается, как правило, в половых клетках, но встречается и в соматических тканях [161, 162]. У млекопитающих пропорции гетероплазмии могут изменяться на более поздних стадиях развития ‒ как следствие неслучайного клеточного или тканеспецифического отбора [161, 163], предположительно не зависящего от клеточной пролиферации [35, 164]. Не следует путать “генетическое бутылочное горлышко”, обозначающее распространение мутации, с понятием “физического бутылочного горлышка” [160, 165] – состоянием, достигаемым физическим уменьшением числа копий молекул мтДНК на клетку во время развития. “Физическое бутылочное горлышко”, в отличие от “генетического бутылочного горлышка”, напрямую соответствует наблюдаемому количеству молекул мтДНК в эксперименте [166]. В отношении существования механизма “генетического бутылочного горлышка” у растений есть разные мнения. Ранее сообщалось о сниженном уровне гетероплазмии у оливковых деревьев, размножавшихся половым путем, в сравнении с деревьями, долгое время размножавшимися вегетативно, что происходит, как предполагается, из-за генетического дрейфа во время формирования женской гаметы [155]. Также есть мнение, что механизм “генетического бутылочного горлышка” в растениях отсутствует, а наблюдаемая гомогенизация последовательностей мтДНК в клетках зародышей растений, ограничивающая распространение мутантных митохондриальных вариантов, обусловлена генной конверсией и эффективной репарацией ошибочного спаривания [167].
В настоящее время недостаточно данных о том, как митохондриальный геном растений с его сложной популяцией молекул ДНК поддерживается и передается от поколения к поколению. Упомянутое ранее массовое слияние митохондрий, наблюдаемое в клетках апикальной меристемы побегов арабидопсиса [68] и в яйцеклетках P. zonale [168], способствует успешному наследованию гетероплазматических геномов благодаря сохранению стабильности митохондриальной популяции в ряду поколений [14, 137]. Тем не менее представленность отдельных последовательностей мтДНК в растительных митохондриях может варьировать в зависимости от ткани/органа растения. Нередко в меристемах и недифференцированных тканях наблюдаются более высокие уровни сублимонов [169‒171]. Показано, что последовательность pvs намного многочисленнее в корневой меристеме, чем в ткани листа фасоли, где она присутствует в виде сублимона [170]. Перестройки хондриома, происходящие во время прорастания семян арабидопсиса, способствуют неравномерному распределению митохондриальных нуклеоидов [85].
Организация митохондриального генома в корневой меристеме риса также существенно отличается от таковой в дифференцированных клетках, что связывают с изменениями стехиометрии молекул мтДНК при ее репликации и распределении во время клеточной дифференцировки [172]. Предполагается, что в меристематической ткани растений находится единая репликационная единица, гарантирующая передачу дочерним клеткам полной генетической информации митохондрий, включая сублимоны [170]. По мере дифференцировки клетки выходят из фазы G1, приобретая обратимое или необратимое состояние покоя (G0) [173]. С течением времени в состоянии необратимого клеточного покоя в зрелых клетках могут накапливаться генетически отличающиеся митохондрии с неравномерным распределением пропорций исходного и мутантного варианта мтДНК.
ВЛИЯНИЕ СТРУКТУРНО-МОРФОЛОГИЧЕСКИХ И ГЕНЕТИЧЕСКИХ ИЗМЕНЕНИЙ НА ФУНКЦИИ МИТОХОНДРИЙ
Влияние гетероплазмии на фенотипы живых организмов
Митохондриальные мутации широко распространены в царстве растений и животных. На животных системах (особенно на людях) митохондриальная гетероплазмия активно изучалась из-за тесной связи между ее возникновением и митохондриальными заболеваниями [174]. Широко известно, что мутации мтДНК у млекопитающих приводят к развитию заболеваний различной степени тяжести. В том случае, когда доля митохондрий с мутантной мтДНК превышает патогенное пороговое значение для ткани, появляется симптоматика заболевания [1]. Одни мутации вызывают нарушения, поражающие нервную систему, мышцы, сердце и эндокринные органы на ранних стадиях развития организма, в то время как другие (чаще всего приобретенные), с более мягкими проявлениями в общих фенотипических чертах, приводят к более позднему началу заболевания [159]. Изменения в мтДНК (вставки, делеции, точечные мутации) могут приводить к респираторным проблемам, эпилепсии, сердечной недостаточности, болезни Паркинсона, диабету, рассеянному склерозу и развитию процесса старения [175‒177]. Однако митохондриальная гетероплазмия клеток животных, включая человека, не всегда приводит к нарушению митохондриальных функций [178‒180]. Многие здоровые люди имеют низкие уровни (<1%) точечных мутаций мтДНК, включая как наследственные, так и приобретенные [159].
У растений особенности структуры и динамики митохондриального генома позволяют поддерживать геномное разнообразие митохондрий без критических воздействий на организм. Первоначально гетероплазмия митохондрий растений также рассматривалась как источник патологических мутаций. Ее обнаруживали в мутантных [181, 182] или гибридных линиях растений [183], а также в растениях, регенерированных из тканевой культуры [184, 185]. Однако позже гетероплазмия была выявлена и в растениях дикого типа [170, 186] и в настоящее время считается нормальным физиологическим состоянием для митохондриальной популяции в растительной клетке [137].
Фенотипические особенности мутантных или гибридных линий растений, обладающих гетероплазмией митохондриального генома, достаточно хорошо изучены. Когда мутации приводят к появлению растений с дефектом, наследуемым по материнской линии, или с мужской стерильностью, их легко обнаружить. Линии, несущие мутации жизненно важных митохондриальных генов в гетероплазматичном состоянии, часто характеризуются замедленным ростом, укорочением стебля, редуцированной и складчатой формой листовой пластины, тогда как гибридные растения, предназначенные исключительно для удовлетворения сельскохозяйственных нужд человека, имеют ценные агрономические характеристики.
Несмотря на вероятную распространенность гетероплазмии в природе [137], пока очень мало известно о ее влиянии на растительные фенотипы в природных популяциях [118]. Альтернативные типы митохондриального генома растений природного происхождения часто отличаются только порядком расположения последовательностей и не содержат мутаций, вредных или существенно влияющих на фенотип [171, 187]. Учитывая множественный механизм возникновения гетероплазмии в клетке, включающий гомологичную рекомбинацию, точечные мутации и двуродительское наследование митохондрий, это явление можно рассматривать как состояние, усложняющее и без того многоуровневую организацию митохондриального генома растений.
Вопрос “нерациональности” организации растительного митохондриального генома подробно обсужден в обзоре M. Woloszynska [137]. Действительно, сложно понять, для чего растениям природных популяций поддерживать гетероплазматическое состояние митохондрий, не дающее им важных фенотипических преимуществ, повышающих их устойчивость и жизнеспособность. Основываясь на универсальности механизмов возникновения гетероплазмии для эукариот, а также на факте существования пластидной гетероплазмии, J. Frey и соавт. [188] высказали предположение, что гетероплазматическое состояние геномов возникло неслучайно и существует в качестве механизма, генерирующего собственное генетическое разнообразие, компенсируя тем самым отсутствие половой рекомбинации. Однако, поскольку к настоящему времени гетероплазмия обнаружена в популяциях лишь нескольких видов растений [189, 190], спешить с выводами относительно ее роли для растительного организма, особенно в отношении фенотипических проявлений, не следует. Необходимы дополнительные исследования распространенности явления митохондриальной гетероплазмии в природных популяциях, что позволит понять роль гетерогенного состояния мтДНК для растительной клетки.
Динамичные изменения структуры хондриома и его биоэнергетические функции
В здоровых, молодых и непролиферирующих (G0) клетках функционально активные митохондрии под воздействием таких факторов, как стресс, изменение потенциала митохондриальных мембран, совместная работа с другими клеточными компартментами, вовлекаются в динамические процессы, влияющие на качество и структурно-морфологические вариации митохондриальной популяции.
Изменения биоэнергетического состояния клетки оказывают влияние на баланс между процессами слияния и деления митохондрий, что, как следствие, приводит к появлению морфологических вариаций [191]. Показано, что высокая активность дыхательной цепи часто коррелирует с удлинением митохондрий. У дрожжей увеличение активности окислительного фосфорилирования сопровождается элонгацией митохондриальной сети [192]. Индукция гиперслияния митохондрий в клетках животных, происходящая при стрессе, также приводит к повышенной выработке митохондриального ATP и определяет стрессоустойчивость клеток [193].
В противоположность этому, высокий энергетический статус в клетках фотосинтезирующих растений обеспечивается преимущественно мелкими дискретными митохондриями. Высокоэнергетическое состояние растительных клеток в дневное время связано с формированием и поддержанием популяции митохондрий небольшого размера, а сниженный энергетический статус в ночное время – с более удлиненными митохондриями [56]. В то же время механизмы энергизации клеток в состоянии клеточной пролиферации универсальны для всех эукариот (рис. 3): непрерывная гиперсеть, формирующаяся на стадии G1-S клеточного цикла, имеет наибольший выход ATP [63, 64, 194, 195], необходимого клетке во время деления, и позволяет распределять энергию на значительные расстояния по всей клетке.
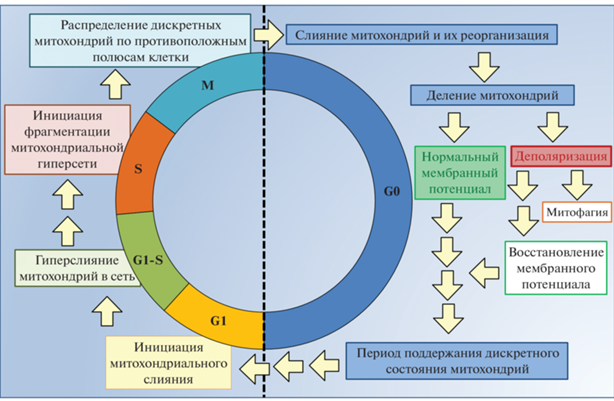
Рис. 3.
Митохондриальная динамика и клеточный цикл. В период клеточной пролиферации происходит последовательная смена этапов преобразования структуры митохондриальной популяции (G1, G1-S, S), заканчивающаяся митозом (M). В фазе клеточного покоя G0 этап кратковременного слияния вызывает деление. После деления дочерняя митохондрия либо поддерживает нормальный мембранный потенциал, либо деполяризуется. Деполяризованные митохондрии могут восстанавливать свой мембранный потенциал и входить в стадию поддержания клеткой состояния дискретных органелл, при этом митохондрии, не восстановившие потенциал по истечении нескольких часов, как правило, удаляются из клетки путем митофагии. При условиях, благоприятствующих вхождению клетки в новый клеточный цикл, дискретные митохондрии снова подвергаются слиянию в фазе G1.
Морфологические изменения митохондрий играют решающую роль в биоэнергетической реактивации при прорастании семян [85]. Покоящиеся сухие семена содержат дискретные небольшие протомитохондрии, после набухания семян митохондрии сливаются, образуя сетчатую структуру, что приводит к биоэнергетической и метаболической реактивации. Временное слияние митохондрий также наблюдается в протопластах непосредственно перед началом деления клеток и в клетках при низких уровнях света, кислорода или сахарозы [56, 69, 196]. Следовательно, образование длинных слитых митохондрий в зиготе, семенах, меристематических клетках способствует активации биоэнергетических процессов для инициации онтогенеза.
В случае повреждений митохондрии, как правило, происходит деполяризация органелл, что ведет к потере потенциала митохондриальной мембраны, необходимого для генерации АТР [197]. Патологические условия также вызывают снижение потенциала митохондриальной мембраны в клетках млекопитающих, при этом часть митохондрий сохраняет свой мембранный потенциал неизменным, что приводит к увеличению гетерогенности митохондриальной популяции [198]. Деполяризованные органеллы различной формы, не подлежащие восстановлению, остаются за пределами пула активных митохондрий, вновь сливающихся в протяженные кластеры [120]. Фрагментированные или неслившиеся митохондрии, не способные поддерживать необходимый уровень синтеза АТР [1, 199], инициируют процессы митофагии, то есть аутофагической деградации митохондрий, и удаляются из клетки (рис. 4). При изучении динамики митохондрий фибробластов показано, что каждая пятая дочерняя митохондрия деполяризуется и элиминируется митофагией [119]. Мелкие фрагментированные митохондрии с выявленной дисфункцией легко захватываются аутофагическими мембранами и затем сливаются с лизосомой [119]. У растений деградация поврежденных или продуцирующих избыток активных форм кислорода митохондрий происходит в вакуоли [200]. Таким образом, динамика митохондрий служит фильтром для отделения и деградации биоэнергетически дисфункциональных митохондрий от здоровых, а митохондриальное деление ‒ первый шаг в поддержании качества митохондриальной популяции.
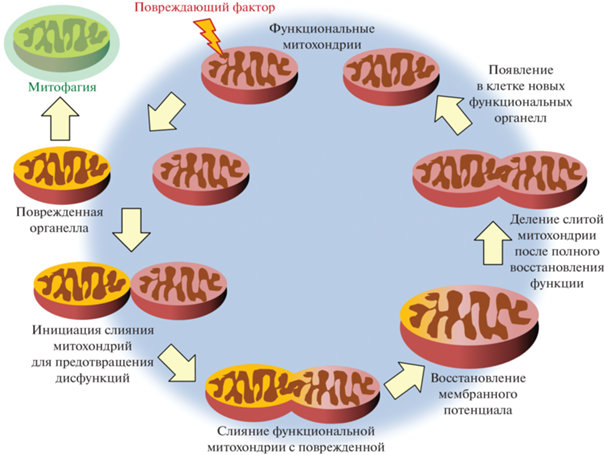
Рис. 4.
Комплементация митохондриальной функции с помощью слияния. Слияние позволяет функциональным митохондриям восстанавливать поврежденные органеллы; при этом происходит распределение компонентов между органеллами и предотвращение неблагоприятных эффектов, вызванных воздействием стрессовых факторов. При альтернативном сценарии поврежденные митохондрии отделяются от здоровой популяции и удаляются из цитоплазмы с помощью аутофагической деградации.
Способность сети образовывать в результате деления отдельные деполяризованные дочерние митохондрии, составляющие преаутофагический пул клетки, считается одной из важных причин возникновения субклеточной гетерогенности в клетках животных [198, 201, 202]. Причина этого, по-видимому, заключается в том, что определенная часть деполяризованных митохондрий не подвергается деградации. Известно, что некоторые митохондрии с дисфункцией производства энергии могут восстанавливать свою поляризацию, стимулируя процессы слияния с пулом активных, высокоэнергетических органелл с последующим разделением [55] (рис. 4).
Ранее была высказана гипотеза, что в пределах единичной клетки одни митохондрии могут специализироваться на хранении наследственной информации, а другие выполнять биоэнергетическую функцию [23]. Но действительно ли некоторая часть деполяризованных митохондрий представляет отдельную субпопуляцию клетки с определенной функцией или они составляют некую популяцию “пула ожидания” для восстановления своей энергетической функциональности, предстоит установить в дальнейших исследованиях.
Биогенез мембран митохондрий и их гетерогенность
На поздних стадиях прорастания образовавшаяся вблизи ядра перинуклеарная тубулоретикулярная митохондриальная структура может способствовать обмену молекулами во всем хондриоме, при этом протомитохондрии становятся своего рода каркасами для встраивания новых полипептидов и липидов [85]. Такие динамичные преобразования хондриома на ранних стадиях развития растения способствуют перекрестному взаимодействию между митохондриальным и ядерным геномами и синхронизации митохондриального биогенеза за счет эффективной доставки и импорта тРНК и полипептидов, кодируемых ядерными транскриптами. Эти изменения сопровождаются усилением транскрипции генов, кодирующих компоненты аппарата импорта внутренней мембраны, фолдинга белков и метаболизма мтДНК [203]. Как показано на культивируемых клетках млекопитающих [87], митохондрии с перинуклеарной локализацией обогащены компонентами аппарата импорта белков внешней мембраны по сравнению с митохондриями на периферии. После окончания прорастания перинуклеарные тубулоретикулярные структуры фрагментируются, что вновь приводит к возникновению популяции физически дискретных митохондрий. Мы предполагаем, что популяция митохондрий уже на стадии молодых проростков имеет различия в белково-липидном составе мембран митохондрий. Это предположение подтверждено результатами наших исследований в области митохондриального импорта ДНК растений in organello [7]. Так, показано, что митохондриальная популяция, выделенная как из молодых проростков кукурузы, так и из запасающей паренхимы репы и 4-недельных листьев арабидопсиса, представляет собой смесь неоднородных органелл и разделяется как минимум на две субфракции [7]. Эти митохондрии отличались по ультраструктуре, дыхательной активности и эффективности импорта ДНК [204] и имели, по-видимому, разный белково-липидный состав мембран. Нами обнаружена высокая эффективность импорта нуклеиновых кислот в одну из митохондриальных субфракций, что может быть обусловлено повышенным уровнем биогенеза мембран в протомитохондриях по сравнению с более зрелыми органеллами. Пока неясно, с чем связана такая гетерогенность: отражает она внутриклеточные процессы или различия в метаболическом статусе отдельных клеток одного типа ткани, а значит и их митохондрий. Ранее в листьях арабидопсиса выявлена гетерогенность митохондриальной структуры при акклиматизации растения к холоду [205]. Также показано [26], что митохондрии, выделенные из молодых листьев гороха, имеют меньшую плотность за счет сниженного содержания некоторых матриксных белков в сравнении с органеллами, изолированными из более старых листьев. На основании этих результатов можно предположить, что листья разного возраста служат источником гетерогенных по молекулярному составу митохондрий.
ЗАКЛЮЧЕНИЕ И ПЕРСПЕКТИВЫ ДАЛЬНЕЙШИХ ИССЛЕДОВАНИЙ
Таким образом, создание и поддержание разнообразия популяции митохондрий растений происходит благодаря особенностям динамики растительных митохондрий. Эта динамика приводит к формированию дискретной организации хондриома с морфологически и генетически разнообразными органеллами, которые могут нести альтернативные варианты митохондриального генома, иметь неравное распределение мтДНК, индивидуальный состав мембран и разную степень митохондриальной энергизации. Согласно некоторым исследованиям, митохондрии, по крайней мере в некоторых типах клеток, могут быть разделены на функционально разнообразные субпопуляции, выполняющие различные задачи в клетке [120]. К важнейшим функциям митохондрий относится биосинтез АТР и поддержание стабильности собственного генома. Однако синтез АТР сопровождается образованием в дыхательной цепи активных форм кислорода, которые, как известно, могут вызывать повреждения в ДНК. Так, Logan [23, 62] считает, что возникает своего рода конфликт в осуществлении митохондриями двух вышеназванных функций. Разрешение этого конфликта он видит в существовании в клетке двух субпопуляций митохондрий, одна из которых выполняет преимущественно энергетическую функцию, а другая служит своего рода генетическим хранилищем. Митохондрии этих субпопуляций могут различаться по содержанию мтДНК и мембранных компонентов, обеспечивающих окислительное фосфорилирование.
Стресс также может вызывать возникновение в клетке специализированных субпопуляций митохондрий. Показано, что при воздействии на лист арабидопсиса патогенными грибами появляется вторая субпопуляция органелл, наряду с типичными митохондриями. В мембране этих органелл происходит накопление белка PEN2, способного формировать гомоолигомерные комплексы [6].
Специализация митохондрий может быть связана с межорганельными взаимодействиями [62] либо характеристиками самой митохондрии, такими как белково-липидный состав мембраны и ее проводимость, а также содержание и состав мтДНК. Различия в белково-липидном составе двойной мембраны митохондрий и, как следствие, в эффективности митохондриального импорта [206] могут служить непрямым доказательством наличия у митохондрий функциональной специализации [7].
Неравномерное распределение нуклеоидов мтДНК среди митохондрий [60, 85] также указывает на потенциальное разделение функций между физически дискретными членами хондриома. Возникновение гетерогенности наблюдается уже в прорастающих семенах, предшествуя усилению митохондриальной динамики и дыхательной активности. Одновременно с этой гипотетической фазой специализации митохондриальных функций происходит рассинхронизация репликации мтДНК и ядерного генома и ослабление ядерного контроля над рекомбинацией мтДНК [85]. Таким образом, есть данные в пользу возможного разделения функций даже внутри протомитохондриальной популяции.
Для дальнейшего понимания природы гетерогенности митохондрий необходимо найти ответы на ряд важных нерешенных вопросов.
1. Анализ корреляции гетероплазмии растительных видов с их фенотипом
На данном этапе мало что известно о роли неоднородного содержания и состояния мтДНК в природных популяциях растений. Для решения этой задачи необходим широкомасштабный мониторинг митогеномов, представляющий особый интерес для исследования филогеографии видов с обширным ареалом. Митохондриальная гетероплазмия имеет важные последствия для эволюции ЦМС растений [207‒209]. Стехиометрия, присущая митохондриальному геному растений, контролируется ядерными генами, поэтому гетероплазмия может быть полезна для установления функционального взаимодействия между двумя геномами.
2. Механизм “поцелуй-и-беги” и функциональная специализация
Дискретный тип организации митохондриальной популяции растений представляет больше возможностей для возникновения специализации у митохондрий. Распространенный механизм “поцелуй-и-беги” ограничивает гомогенизацию содержимого митохондрий, будучи источником фенотипической и генотипической митохондриальной гетерогенности. Однако вопрос, каким образом реализуется последующая специализация митохондрий, остается открытым. Возможно, митохондрии в короткий срок до следующего временного слияния становятся в некоторой степени специализированными или по мере прохождения нескольких циклов слияния-деления постепенно приобретают специализацию. Сохранение таких параметров, как индивидуальная морфология, состав внутренней митохондриальной мембраны и ассоциированной с ней мтДНК, которые были свойственны данной митохондрии до временного слияния [94, 96], свидетельствует о возможном сохранении и ее специализации.
3. Деполяризованные митохондрии: функция или дисфункция
В настоящий момент не до конца выяснено, действительно ли некоторая часть деполяризованных митохондрий составляет отдельную субпопуляцию клетки с определенной функцией или же это “пул ожидания” для восстановления своей энергетической функциональности либо деградации.
4. Различия в способности к импорту у субклеточных популяций митохондрий
Ранее нами выявлена гетерогенность митохондрий в клетках различных растительных тканей [7]. Оказалось, что отдельные субфракции митохондрий, выделенные из листьев арабидопсиса, паренхимы корнеплодов репы и меристематической ткани колеоптилей кукурузы обладают различающимися структурными и функциональными характеристиками. Особо интригующим результатом оказалась различная способность выделенных митохондриальных субфракций к импорту ДНК. Является ли источником этих различий внутриклеточная гетерогенность митохондриальной популяции, или же мы имеем дело с не менее интересным феноменом межклеточной гетерогенности в пределах одного типа ткани? Ответ на этот вопрос предстоит получить в дальнейших исследованиях.
В работе использовано оборудование Центра Коллективного Пользования “Биоаналитика” Сибирского института физиологии и биохимии растений Сибирского отделения Российской академии наук (г. Иркутск).
Работа выполнена при финансовой поддержке Российского фонда фундаментальных исследований в рамках научного проекта № 20-14-50001.
Настоящая статья не содержит описания выполненных авторами исследований с участием людей или использованием животных в качестве объектов.
Авторы заявляют об отсутствии конфликта интересов.
Список литературы
Kornick K., Bogner B., Sutter L., Das M. (2019) Population dynamics of mitochondria in cells: a mini-mal mathematical model. Front. Phys. 7, 146. https://doi.org/10.3389/fphy.2019.00146
Welchen E., Garciìa L., Mansilla N., Gonzalez D.H. (2014) Coordination of plant mitochondrial biogenesis: keeping pace with cellular requirements. Front. Plant Sci. 4, 551. https://doi.org/10.3389/fpls.2013.00551
Galloway C.A., Yoon Y. (2013) Mitochondrial morphology in metabolic diseases. Antioxid. Redox Signal. 19, 415–430. https://doi.org/10.1089/ars.2012.4779
Mishra N.C., Kumar S. (2005) Apoptosis: a mitochondrial perspective on cell deathю Indian J. Exp. Biol. 43, 25–34.
Howell K.A., Millar A.H., Whelan J. (2006) Ordered assembly of mitochondria during rice germination begins with promitochondrial structures rich in components of the protein import apparatus. Plant Mol. Biol. 60, 201–223. https://doi.org/10.1007/s11103-005-3688-7
Fuchs R., Kopischke M., Klapprodt C., Hause G., Meye A.J., Schwarzländer M., Fricker M.D., Lipka V. (2016) Immobilized subpopulations of leaf epidermal mitochondria mediate PENETRATION2-dependent pathogen entry control in Arabidopsis. Plant Cell. 28, 130–145. https://doi.org/10.1105/tpc.15.00887
Tarasenko T.A., Subota I.Yu., Tarasenko V.I., Konstantinov Y.M., Koulintchenko M.V. (2020) Plant mitochondrial subfractions have different ability to import DNA. Theor. Exp. Plant Physiol. 32, 5–18. https://doi.org/10.1007/s40626-020-00167-w
Murcha M.W., Huang T., Whelan J. (1999) Import of precursor proteins into mitochondria from soybean tissues during development. FEBS Lett. 464, 53‒59. https://doi.org/10.1016/s0014-5793(99)01674-9
Weber-Lotfi F., Koulintchenko M., Ibrahim N., Hammann P., Mileshina D., Konstantinov Yu.M., Dietrich A. (2015) Nucleic acid import into mitochondria: new insights into the translocation pathways. Biochim. Biophys. Acta. 1853, 3165–3181. https://doi.org/10.1016/j.bbamcr.2015.09.011
Millar A.H., Sweetlove L.J., Giegé P., Leaver C.J. (2001) Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol. 127, 1711–1727.
Stickens D., Verbelen J.P. (1996) Spatial structure of mitochondria and ER denotes changes in cell physiology of cultured tobacco protoplast. Plant. J. 9, 85–92. https://doi.org/10.1046/j.1365-313X.1996.09010085.x
Logan D.C., Leaver C.J. (2000) Mitochondria-targeted GFP highlights the heterogeneity of mitochondrial shape, size and movement within living plant cells. J. Exp. Bot. 51, 865–871. https://doi.org/10.1093/jexbot/51.346.865
Wai T., Langer T. (2016) Mitochondrial dynamics and metabolic regulation. Trends Endocrinol. Metab. 27, 105–117. https://doi.org/10.1016/j.tem.2015.12.001
Johnston I.G. (2019a) Tension and resolution: dynamic, evolving populations of organelle genomes within plant cells. Mol. Plant. 12, 764–783. https://doi.org/10.1016/j.molp.2018.11.002
Frisell W.R., Patwardhan M.V., Mackenzie C.G. (1965) Quantitative studies on the soluble compartments of light and heavy mitochondria from rat liver. J. Biol. Chem. 240, 1829‒1835.
Graham J.M. (2002) OptiPrep density gradient solutions for nonmammalian organelles. ScientificWorldJournal. 2, 1444–1448. https://doi.org/10.1100/tsw.2002.839
Белякович А.Г. (1990) Изучение митохондрий и бактерий c помощью соли тетразолия п-НТФ. Пущино: ОНТИ НЦБИ.
Шишмаков Д.А., Анисимов P.Л., Векшин Н.Л. (2004) Некоторые свойства протомитохондрий. Биол. мембраны. 21, 389‒395.
Dai H., Lo Y.S., Jane W.N., Lee L.W., Chiang K.S. (1998) Population heterogeneity of higher-plant mitochondria in structure and function. Eur. J. Cell Biol. 75, 198‒209. https://doi.org/10.1016/S0171-9335(98)80062-9
Logan D.C., Millar A.H., Sweetlove L.J., Hill S.A., Leaver C.J. (2001) Mitochondrial biogenesis during germination in maize embryos. Plant Physiol. 125, 662‒672. https://doi.org/10.1104/pp.125.2.662
Begunova E.A., Vekshin N.L. (2015) Protomitohondria from liver cells: similarities with and differences from mitochondria. Biophysics. 60, 921–927. https://doi.org/10.1134/S0006350915060032
Lund H.A., Vatter A.E., Hanson J.B. (1958) Biochemical and cytological changes accompanying growth and differentiation in the roots of Zea mays. J. Biophis. Biochem. Cytol. 4, 87‒96. https://doi.org/10.1083/jcb.4.1.87
Logan D.C. (2006) The mitochondrial compartment. J. Exp. Bot. 57, 1225–1243. https://doi.org/10.1093/jxb/erj151
Petrussa E., Bertolini A., Krajnáková J., Casolo V., Macrì F., Vianello A. (2008) Isolation of mitochondria from embryogenic cultures of Picea abies (L.) Karst. and Abies cephalonica Loud.: characterization of a K+ATP channel. Plant Cell Rep. 27, 137‒146. https://doi.org/10.1007/s00299-007-0436-2
Bakeeva L.E., Kirnos M.D., Aleksandrushkina N.I., Kazimirchyuk S.B., Shorning B.Yu., Zamyatnina V.A., Yaguzhinsky L.S., Vanyushin B.F. (1999) Subcellular reorganization of mitochondria producing heavy DNA in aging wheat coleoptiles. FEBS Lett. 457, 122‒125. https://doi.org/10.1016/s0014-5793(99)01025-x
Vauclare P., Diallo N., Bourguignon J., Macherel D., Douce R. (1996) Regulation of the expression of the glycine decarboxylase complex during pea leaf development. Plant Physiol. 112, 1523–1530. https://doi.org/10.1104/pp.112.4.1523
des Francs-Small C.C., Ambard-Bretteville F., Darpas A., Sallantin M., Huet J.C., Pernollet J.C., Rémy R. (1992) Variation of the polypeptide composition of mitochondria isolated from different potato tissues. Plant Physiol. 98, 273‒278. https://doi.org/10.1104/pp.98.1.273
Bahl J., Demandre C., Chauveau M., Alpha M.J., Roussaux J. (1997) Lipid changes in mitochondria of Arum maculatum spadix during inflorescence development. Plant Physiol. Biochem. 35, 693‒700. https://doi.org/10.1007/978-94-015-8394-7_55
Small I.D., Isaac P.G., Leaver C.J. (1987) Stoichiometric differences in DNA molecules containing the atpA gene suggest mechanisms for the generation of mitochondrial diversity in maize. EMBO J. 6, 865‒869. https://doi.org/10.1002/j.1460-2075.1987.tb04832.x
Birky C.W. (2001) The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu. Rev. Genet. 35, 125–148. https://doi.org/ 10.1146/annurev.genet.35.102401.090231
Aryaman J., Johnston I.G. Jones N.S. (2018) Mitochondrial heterogeneity. Front. Genet. 9, 718. https://doi.org/10.3389/fgene.2018.00718
Nishimura M., Douce R., Akazawa T. (1982) Isolation and characterization of metabolically competent mitochondria from spinach leaf protoplasts. Plant Physiol. 69, 916‒920. https://doi.org/10.1104/pp.69.4.916
Collins T.J., Berridge M.J., Lipp P., Bootman M.D. (2002) Mitochondria are morphologically and functionally heterogeneous within cells. EMBO J. 21, 1616–1627. https://doi.org/10.1093/emboj/21.7.1616
Simeonova E., Garstka M., Koziol-Lipi’nska J., Mostowska A. (2004) Monitoring the mitochondrial transmembrane potential with the JC-1 fluorochrome in programmed cell death during mesophyll leaf senescence. Protoplasma. 223, 143‒153. https://doi.org/10.1007/s00709-004-0039-5
Battersby B.J., Shoubridge E.A. (2001) Selection of a mtDNA sequence variant in hepatocytes of heteroplasmic mice is not due to differences in respiratory chain function or efficiency of replication. Hum. Mol. Genet. 10, 2469–2479. https://doi.org/10.1093/hmg/10.22.2469
Staehelin L.A. (1997) The plant ER: a dynamic orga-nelle composed of a large number of discrete functional domains. Plant J. 11, 1151‒1165. https://doi.org/10.1046/j.1365-313x.1997.11061151.x
Köhler R.H., Cao J., Zipfel W.R., Webb W.W., Hanson M.R. (1997) Exchange of protein molecules through connections between higher plant plastids. Science. 276, 2039‒2042. https://doi.org/10.1126/science.276.5321.2039
Smart C.J., Monéger F., Leaver C.J. (1994) Cell-specific regulation of gene expression in mitochondria during anther development in sunflower. Plant Cell. 6, 811–825. https://doi.org/10.1105/tpc.6.6.811
Southworth D., Strout G., Russell S.D. (1997) Freeze-fracture of sperm of Plumbago zeylanica L. in pollen and in vitro. Sex. Plant Reprod. 10, 217–226. https://doi.org/10.1007/s004970050090
Ehrenshaft M., Brambl R. (1990) Respiration and mitochondrial biogenesis in germinating embryos of maize. Plant Physiol. 93, 295–304. https://doi.org/10.1104/pp.93.1.295
Botha F.C., Potgieter G.P., Botha A.M. (1992) Respiratory metabolism and gene expression during seed germination. Plant Growth Reg. 11, 211‒224. https://doi.org/10.1007/BF00024560
Bewley J.D. (1997) Seed germination and dormancy. Plant Cell. 9, 1055‒1066. https://doi.org/10.1105/tpc.9.7.1055
Bain M.J., Mercer F.V. (1964) Organization resistance and respiration climacteric. Aust. J. Biol. Sci. 17, 78–85. https://doi.org/10.1071/BI9640078
Malhotra S.S., Spencer M. (1973) Structural development during germination of different populations of mitochondria from pea cotyledons. Plant Physiol. 52, 575–579. https://doi.org/10.1104/pp.52.6.575
Falk K.L., Behal R.H., Xiang C.B., Oliver D.J. (1998) Metabolic bypass of the tricarboxylic acid cycle during lipid mobilization in germinating oilseeds: regulation of NAD+-dependent isocitrate dehydrogenase versus fumarase. Plant Physiol. 117, 473–481. https://doi.org/10.1104/pp.117.2.473
Daley D.O., Considine M.J., Howell K.A., Millar A.H., Day D.A., Whelan J. (2003) Respiratory gene expression in soybean cotyledons during post-germinative development. Plant Mol. Biol. 51, 745–755. https://doi.org/10.1023/a:1022502501373
Nunnari J., Marshall W.F., Straight A., Murray A., Sedat J.W., Walter P. (1997) Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell. 8, 1233-1242. https://doi.org/10.1091/mbc.8.7.1233
Okamoto K., Shaw J.M. (2005) Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 39, 503–536. https://doi.org/10.1146/annurev.genet.38.072902.093019
Arimura S., Tsutsumi N. (2002) A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc. Natl. Acad. Sci. USA. 99, 5727‒5731. https://doi.org/10.1073/pnas.082663299
Bereiter-Hahn J. (1990) Behavior of mitochondria in the living cell. Int. Rev. Cytol. 122, 1‒63. https://doi.org/10.1016/s0074-7696(08)61205-x
Rizzuto R., Pinton P., Carrington W., Fay F.S., Fogarty K.E., Lifshitz L.M., Tuft R.A., Pozzan T. (1998) Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 280, 1763–1766. https://doi.org/10.1126/science.280.5370.1763
Aryaman J., Bowles C., Jones N.S., Johnston I.G. (2019) Mitochondrial network state scales mtDNA genetic dynamics. Genetics. 212, 1429–1443. https://doi.org/10.1534/genetics.119.302423
Xie L.-L., Shi F., Tan Z., Li Y., Bode A.M., Cao Y. (2018) Mitochondrial network structure homeostasis and cell death. Cancer Sci. 109, 3686–3694. https://doi.org/10.1111/cas.13830
Rafelski S.M. (2013) Mitochondrial network morphology: building an integrative, geometrical view. BMC Biology. 11, 71. https://doi.org/10.1186/1741-7007-11-71
Youle R.J., van der Bliek A.M. (2012) Mitochondrial fission, fusion, and stress. Science. 337, 1062–1065. https://doi.org/10.1126/science.1219855
Jaipargas E.-A., Barton K. A., Mathur N., Mathur J. (2015) Mitochondrial pleomorphy in plant cells is driven by contiguous ER dynamics. Front. Plant Sci. 6, 783. https://doi.org/10.3389/fpls.2015.00783
Chen H., Chan D.C. (2005) Emerging functions of mammalian mitochondrial fusion and fission. Hum. Mol. Genet. 14, R283–R289. https://doi.org/10.1093/hmg/ddi270
Fuchs P., Rugen N., Carrie C., Elsässer M., Finkemeier I., Giese J., Hildebrandt T.M., Kühn K., Maurino V.G., Ruberti C., Schallenberg-Rüdinger M., Steinbeck J., Braun H.-P., Eubel H., Meyer E.H., Müller-Schüssele S.J., Schwarzländer M. (2020) Single organelle function and organization as estimated from Arabidopsis mitochondrial proteomics. Plant J. 101, 420–441. https://doi.org/10.1111/tpj.14534
Sheahan M.B., Rose R.J., McCurdy D.W. (2004) Organelle inheritance in plant cell division: the actin cytoskeleton is required for unbiased inheritance of chloroplasts, mitochondria and endoplasmic reticulum in dividing protoplasts. Plant J. 37, 379–390. https://doi.org/10.1046/j.1365-313X.2003.01967.x
Preuten T., Cincu E., Fuchs J., Zoschke R., Liere K., Borner T. (2010) Fewer genes than organelles: extremely low and variable gene copy numbers in mitochondria of somatic plant cells. Plant J. 64, 948–959. https://doi.org/10.1111/j.1365-313X.2010.04389.x
Arimura S., Aida G.P., Fujimoto M., Nakazono M., Tsutsumi N. (2004b) Arabidopsis dynamin-like protein 2a(ADL2a), likeADL2b, is involved in plant mitochondrial division. Plant Cell Physiol. 45, 236–242. https://doi.org/10.1093/pcp/pch024
Logan D.C. (2010) Mitochondrial fusion, division and positioning in plants. Biochem. Soc. Trans. 38, 789–795. https://doi.org/10.1042/BST0380789
Seguí-Simarro J.M., Coronado M.J., Staehelin L.A. (2008) The mitochondrial cycle of Arabidopsis shoot apical meristem and leaf primordium meristematic cells is defined by a perinuclear tentaculate/cage-like mitochondrion. Plant Physiol. 148, 1380–1393. https://doi.org/10.1104/pp.108.126953
Kianian P.M.A., Kianian S.F. (2014) Mitochondrial dynamics and the cell cycle. Front. Plant Sci. 5, 222. https://doi.org/10.3389/fpls.2014.00222
Christiansen E.G. (1949) Orientation of the mitochondria during mitosis. Nature. 163, 361.
Taguchi N., Ishihara N., Jofuku A., Oka T., Mihara K. (2007) Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 282, 11521‒11529. https://doi.org/10.1074/jbc.M607279200
Kanfer G., Courthéoux T., Peterka M., Meier S., Soste M., Melnik A., Reis K., Aspenstrom P., Peter M., Picotti P., Kornmann B. (2015) Mitotic redistribution of the mitochondrial network by Miro and Cenp-F. Nat. Commun. 6, 8015. https://doi.org/10.1038/ncomms9015
Seguí-Simarro J.M., Staehelin L.A. (2009) Mitochondrial reticulation in shoot apical meristem cells of Arabidopsis provides a mechanism for homogenization of mtDNA prior to gamete formation. Plant Signal. Behav. 4, 168–171. https://doi.org/10.4161/psb.4.3.7755
Sheahan M.B., McCurdy D.W., Rose R.J. (2005) Mitochondria as a connected population: ensuring continuity of the mitochondrial genome during plant cell dedifferentiation through massive mitochondrial fusion. Plant J. 44,744–755. https://doi.org/10.1111/j.1365-313X.2005.02561.x
Katajisto P., Döhla J., Chaffer C.L., Pentinmikko N., Marjanovic N., Iqbal S., Zoncu R., Chen W., Weinberg R.A., Sabatini D.M. (2015) Stem cells. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science. 348, 340–343. https://doi.org/10.1126/science.1260384
Kanfer G., Kornmann B. (2016) Dynamics of the mitochondrial network during mitosis. Biochem. Soc. Trans. 44, 510–516. https://doi.org/10.1042/BST20150274
Bird J., Porter E.K., Dickinson H.G. (1983). Events in the cytoplasm during male. J. Cell Sci. 42, 27–42
Mogenson H.L., Rusche L. (1985) Quantitative ultrastructural analysis of barley sperm. I. Occurrence and mechanism of cytoplasm and organelle reduction and the question of sperm dimorphism. Protoplasma. 13, 1–13.https://doi.org/10.1007/BF01273229
McConchie C.A., Hough T., Knox R.B. (1987) Ultrastructural analysis of the sperm cells of mature pollen of maize, Zea mays. Protoplasma. 139, 9–19. https://doi.org/10.1007/BF01417530
Wagner V.T., Dumas C., Mogensen H.L. (1988) Morphometric analysis of isolated Zea mays sperm. J. Cell Sci. 93, 179–184. https://doi.org/10.1242/jcs.93.1.179
Mogensen H.L., Wagner V.T., Dumas C. (1990) Quantitative, three-dimensional ultrastructure of isolated corn (Zea mays) sperm cells. Protoplasma. 153, 136–140. https://doi.org/10.1007/BF01353997
Mahrous E., Yang Q., Clarke H.J. (2012) Regulation of mitochondrial DNA accumulation during oocyte growth and meiotic maturation in the mouse. Reproduction. 144, 177–185. https://doi.org/10.1530/REP-12-0113
Dalton C.M., Carroll J. (2013) Biased inheritance of mitochondria during asymmetric cell division in the mouse oocyte. J. Cell Sci. 126, 2955–2964. https://doi.org/10.1242/jcs.128744
Faure J., Mogensen H.L., Kranz E., Digonnet C., Dumas C. (1992) Ultrastructural characterization and three-dimensional reconstruction of isolated maize (Zea mays L.) egg cell protoplasts. Protoplasma. 171, 97–103. https://doi.org/10.1007/BF01403723
Schulz P., Jensen W.A. (1973) Capsella embryogenesis: the central cell. J. Cell Sci. 12, 741–763.
Kuroiwa H., Ohta T., Kuroiwa T. (1996) Studies on the development and three-dimensional reconstruction of giant. Protoplasma. 192, 235–244. https://doi.org/10.1007/BF01273895
Yamaoka S., Nakajima M., Fujimoto M., Tsutsumi N. (2011) MIRO1 influences the morphology and intracellular distribution of mitochondria during embryo-nic cell division in Arabidopsis. Plant Cell Rep. 30, 239–244. https://doi.org/10.1007/s00299-010-0926-5
Kimata Y., Higaki T., Kurihara D., Ando N., Matsumoto H., Higashiyama T., Ueda M. (2020) Mitochondrial dynamics and segregation during the asymmetric division of Arabidopsis zygotes. Quant. Plant Biol. 1, e3. https://doi.org/10.1017/qpb.2020.4
Gooh K., Ueda M., Aruga K., Park J., Arata H., Higashiyama T., Kurihara D. (2015) Live-cell imaging and optical manipulation of Arabidopsis early embryogenesis. Dev. Cell. 34, 242–251. https://doi.org/10.1016/j.devcel.2015.06.008
Paszkiewicz G., Gualberto J.M., Benamar A., Macherel D., Logan D.C. (2017) Arabidopsis seed mitochondria are bioenergetically active immediately upon imbibition and specialize via biogenesis in preparation for autotrophic growth. Plant Cell. 29, 109–128. https://doi.org/10.1105/tpc.16.00700
Quintana A., Hoth M. (2012) Mitochondrial dyna-mics and their impact on T cell function. Cell Calcium. 52, 57–63. https://doi.org/10.1016/j.ceca.2012.02.005
Wurm C.A., Neumann D., Lauterbach M.A., Harke B., Egner A., Hell S.W., Jakobs S. (2011) Nanoscale distribution of mitochondrial import receptor Tom20 is adjusted to cellular conditions and exhibits an inner-cellular gradient. Proc. Natl. Acad. Sci. USA. 108, 13546–13551. https://doi.org/10.1073/pnas.1107553108
Smirnova E., Griparic L., Shurland D.L., van der Bliek A.M. (2001) Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 12, 2245‒2256. https://doi.org/10.1091/mbc.12.8.2245
van der Bliek A.M., Shen Q., Kawajiri S. (2013) Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 5, a011072. https://doi.org/10.1101/cshperspect.a011072
Chang C.R., Blackstone C. (2010) Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann. NY Acad. Sci. 1201, 34‒39. https://doi.org/10.1111/j.1749-6632.2010.05629.x
Park Y.-Y., Cho H. (2012) Mitofusin 1 is degraded at G2/M phase through ubiquitylation by MARCH5. Cell Div. 7, 25. https://doi.org/10.1186/1747-1028-7-25
Chen H., Chomyn A., Chan D.C. (2005) Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 280, 26185–26192. https://doi.org/10.1074/jbc.M503062200
Logan D.C., Scott I., Tobin A.K. (2003) The genetic control of plant mitochondrial morphology and dynamics. Plant J. 36, 500–509. https://doi.org/10.1046/j.1365-313x.2003.01894.x
Arimura S., Fujimoto M., Doniwa Y., Kadoya N., Nakazono M., Sakamoto W. Tsutsumi N. (2008) Arabidopsis elongated mitochondria1 is required for loca-lization of dynamin-related protein3A to mitochondrial fission sites. Plant Cell. 20, 1555–1566. https://doi.org/10.1105/tpc.108.058578
El Zawily A.M., Schwarzlander M., Finkemeier I., Johnston I.G., Benamar A., Cao Y., Gissot C., Meyer A.J., Wilson K., Datla R., Macherel D., Jones N.S., Logan D.C. (2014) Friendly regulates mitochondrial distribution, fusion, and quality control in Arabidopsis. Plant Physiol. 166, 808–828. https://doi.org/10.1104/pp.114.243824
Hong. Z., Bednarek. S.Y., Blumwald. E., Hwang I., Jurgens G., Menzel D., Osteryoung K.W., Raikhel N.V., Shinozaki K., Tsutsumi N., Verma D.P. (2003) A unified nomenclature for Arabidopsis dynamin-related large GTPases based on homology and possible functions. Plant Mol. Biol. 53, 261–265. https://doi.org/10.1023/b:plan.0000007000.29697.81
Arimura S.I., Yamamoto J., Paul Aida G., Nakazono M., Tsutsumi N. (2004a) Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc. Natl. Acad. Sci. USA. 101, 7805–7808. https://doi.org/10.1073/pnas.0401077101
Logan D.C., Scott I., Tobin A.K. (2004) ADL2a, like ADL2b, is involved in the control of higher plant mitochondrial morphology. J. Exp. Bot. 55, 783–785. https://doi.org/10.1093/jxb/erh073
Aung K., Hu J. (2012) Differential roles of Arabidopsis dynamin-related proteins DRP3A, DRP3B, and DRP5B in organelle division. J. Integr. Plant Biol. 54, 921–931. https://doi.org/10.1111/j.1744-7909.2012.01174.x
Scott I., Tobin A.K., Logan D.C. (2006) BIGYIN, an orthologue of human and yeast FIS1 genes functions in the control of mitochondrial size and number in Arabidopsis thaliana. J. Exp. Bot. 57, 1275–1280. https://doi.org/10.1093/jxb/erj096
Arimura S.I. (2018) Fission and fusion of plant mitochondria, and genome maintenance. Plant Physiol. 176, 152–161. https://doi.org/10.1104/pp.17.01025
Pan R., Hu J. (2015) Plant mitochondrial dynamics and the role of membrane lipids. Plant Signal. Behav. 10, e1050573. https://doi.org/10.1080/15592324.2015.1050573
Zhang X.C., Hu J.P. (2008) FISSION1A and FISSION1B proteins mediate the fission of peroxisomes and mitochondria in Arabidopsis. Mol. Plant. 1, 1036–1047. https://doi.org/10.1093/mp/ssn056
Aung K., Hu J. (2011) The Arabidopsis tail-anchored protein peroxisomal and mitochondrial division factor1 is involved in the morphogenesis and proliferation of peroxisomes and mitochondria. Plant Cell. 23, 4446–4461. https://doi.org/10.1105/tpc.111.090142
Rose R.J. (2021) Contribution of massive mitochondrial fusion and subsequent fission in the plant life cycle to the integrity of the mitochondrion and its genome. Int. J. Mol. Sci. 22, 5429. https://doi.org/10.3390/ijms22115429
White R.R., Lin C., Leaves I., Castro I.G., Metz J., Bateman B.C., Botchway S.W., Ward A.D., Ashwin P., Sparkes I. (2020) Miro2 tethers the ER to mitochondria to promote mitochondrial fusion in tobacco leaf epidermal cells. Commun. Biol. 3, 161. https://doi.org/10.1038/s42003-020-0872-x
Altmann K., Frank M., Neumann D., Jakobs S. Westermann B. (2008) The class V myosin motor protein, Myo2, plays a major role in mitochondrial motility in Saccharomyces cerevisiae. J. Cell Biol. 181, 119–130. https://doi.org/10.1083/jcb.200709099
Varadi A., Johnson-Cadwell L.I., Cirulli V., Yoon Y., Allan V.J. Rutter G.A. (2004) Cytoplasmic dynein regulates the subcellular distribution of mitochondria by controlling the recruitment of the fission factor dynamin-related protein-1. J. Cell Sci. 117, 4389–4400. https://doi.org/10.1242/jcs.01299
Rattner J.B., Rao A., Fritzle, M.J., Valencia D.W., Yen T.J. (1993) CENP-F is a ca 400 kDa kinetochore protein that exhibits a cell-cycle dependent localization. Cell Motil. Cytoskeleton. 26, 214–226. https://doi.org/10.1002/cm.970260305
Lackner L.L., Ping H., Graef M., Murley A., Nunnari J. (2013) Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc. Natl. Acad. Sci. USA. 110, E458–E467. https://doi.org/10.1073/pnas.1215232110
Rizzuto R., Brini M., Murgia M., Pozzan T. (1993) Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science. 262, 744–747. https://doi.org/10.1126/science.8235595
Achleitner G., Gaigg B., Krasser A., Kainersdorfer E., Kohlwein S.D., Perktold A., Zellnig G., Daum G. (1999) Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorga-nelle transport of phospholipids through membrane contact. Eur. J. Biochem. 264, 545–553. https://doi.org/10.1046/j.1432-1327.1999.00658.x
Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., Voeltz G.K. (2011) ER tubules mark sites of mitochondrial division. Science. 334, 358–362. https://doi.org/10.1126/science.1207385
Mootha V.K., Bunkenborg J., Olsen J.V., Hjerrild M., Wisniewski J.R., Stahl E., Bolouri M.S., Ray H.N., Sihag S., Kamal M., Patterson N., Lander E.S., Mann M. (2003) Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 115, 629–640. https://doi.org/10.1016/s0092-8674(03)00926-7
Robertson E.J., Williams M., Harwood J.L., Lindsay J.G., Leaver C.J., Leech R.M. (1995) Mitochondria increase three-fold and mitochondrial proteins and lipid change dramatically in postmeristematic cells in young wheat leaves grown in elevated CO2. Plant Physiol. 108, 469-474. https://doi.org/10.1104/pp.108.2.469
Sakamoto W., Takami T. (2018) Chloroplast DNA dynamics: copy number, quality control and degradation. Plant Cell Physiol. 59, 1120–1127. https://doi.org/10.1093/pcp/pcy084
Kmiec B., Woloszynska M., Janska H. (2006) Heteroplasmy as a common state of mitochondrial genetic information in plants and animals. Curr. Genet. 50, 149-159. https://doi.org/10.1007/s00294-006-0082-1
Levsen N., Bergero R., Charlesworth D., Wolff K. (2016) Frequent, geographically structured heteroplasmy in the mitochondria of a flowering plant, ribwort plantain (Plantago lanceolata). Heredity. 117, 1–7. https://doi.org/10.1038/hdy.2016.15
Twig G., Elorza A., Molina A.J.A., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., Alroy J., Wu M., Py B.F., Yuan J., Deeney J.T., Corkey B.E., Shirihai O.S. (2008) Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27, 433–446. https://doi.org/10.1038/sj.emboj.7601963
Wikstrom J.D., Twig G., Shirihai O.S. (2009) What can mitochondrial heterogeneity tell us about mitochondrial dynamics and autophagy? Int. J. Biochem. Cell Biol. 41, 1914–1927. https://doi.org/10.1016/j.biocel.2009.06.006
Sukhorukov V.M., Dikov D., Busch K., Strecker V., Wittig I., Bereiter-Hahn J. (2010) Determination of protein mobility in mitochondrial membranes of living cells. Biochim. Biophys. Acta. 1798, 2022–2032. https://doi.org/10.1016/j.bbamem.2010.07.016
Wallace D.C. (2007) Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu. Rev. Biochem. 76, 781‒821. https://doi.org/10.1146/annurev.biochem.76.081205.150955
Jakobs S., Stoldt S., Neumann D. (2011) Light microscopic analysis of mitochondria heterogeneity in cell population and within single cells. Adv. Biochem. Eng. Biotechnol. 123, 1–19. https://doi.org/10.1007/10_2010_81
Morley S.A., Nielsen B.L. (2017) Plant mitochondrial DNA. Front. Biosci. 22, 1023‒1032. https://doi.org/10.2741/4531
Szczepanowska J., Malinska D., Wieckowski M.R., Duszynski J. (2012) Effect of mtDNA point mutations on cellular bioenergetics. Biochim. Biophys. Acta. 1817, 1740‒1746. https://doi.org/10.1016/j.bbabio.2012.02.028
Wallace D.C., Fan W. (2009) The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev. 23, 1714‒1736. https://doi.org/10.1101/gad.1784909
Lee S., Kim S., Sun X., Lee J.H., Cho H. (2007) Cell cycle-dependent mitochondrial biogenesis and dynamics in mammalian cells. Biochem. Biophys. Res. Commun. 357, 111–117. https://doi.org/10.1016/j.bbrc.2007.03.091
Parone P.A., Da Cruz S., Tondera D., Mattenberger Y., James D.I., Maechler P., Barja F., Martinou J.-C. (2008) Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS One. 3, e3257. https://doi.org/10.1371/journal.pone.0003257
Yoon Y.S., Yoon D.S., Lim I.K., Yoon S.H., Chung H.Y., Rojo M., Malka F., Jou M.J., Martinou J.C., Yoon G. (2006) Formation of elongated giant mitochondria in DFO-induced cellular senescence: involvement of enhanced fusion process through modulation of Fis1. J. Cell Physiol. 209, 468–480. https://doi.org/10.1002/jcp.20753
Tateda C., Watanabe K., Kusano T., Takahashi Y. (2011) Molecular and genetic characterization of the gene family encoding the voltage-dependent anion channel in Arabidopsis. J. Exp. Bot. 62, 4773–4785. https://doi.org/10.1093/jxb/err113
Rice D.W., Alverson A.J., Richardson A.O., Young G.J., Sanchez-Puerta M.V., Munzinger J., Barry K., Boo-re J.L., Yan Zhang Y., De Pamphilis C.W., Knox E.B., Palmer J.D. (2013) Horizontal transfer of entire genomesvia mitochondrial fusion in the angiosperm Amborella. Science. 342, 1468–1473. https://doi.org/10.1126/science.1246275
Mower J.P., Sloan D.B., Alverson A.J. (2012) Plant mitochondrial genome diversity: the genomics revolution. Plant Gen. Div. 1, 123–144. https://doi.org/10.1007/978-3-7091-1130-7_9
Lonsdale D.M., Brears T., Hodge T.P., Melville S.E., Rottmann W.H. (1988) The plant mitochondrial genome: homologous recombination as mechanism for generating heterogeneity. Philos. Trans. 319, 149–163. https://doi.org/10.1098/rstb.1988.0039
Legros F., Malka F., Frachon P., Lombès A., Rojo M. (2004) Organization and dynamics of human mitochondrial DNA. J. Cell Sci. 117, 2653–2662. https://doi.org/10.1242/jcs.01134
Arciuch V.G.A., Elguero M.E., Poderoso J.J., Carreras M.C. (2012) Mitochondrial regulation of cell cycle and proliferation. Antioxid. Redox Signal. 16, 1150–1180. https://doi.org/10.1089/ars.2011.4085
Gualberto J.M., Mileshina D., Wallet C., Niazi A.K., Weber-Lotfi F., Dietrich A. (2014) The plant mitochondrial genome: dynamics and maintenance. Biochimie. 100, 107–120. https://doi.org/10.1016/j.biochi.2013.09.016
Woloszynska M. (2010) Heteroplasmy and stoichiometric complexity of plant mitochondrial genomes—though this be madness, yet there’s method in’t. J. Exp. Bot. 61, 657–671, https://doi.org/10.1093/jxb/erp361
Abdelnoor R.V., Yule R., Elo, A., Christensen A.C., Meyer-Gauen G., Mackenzie S.A. (2003) Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc. Natl. Acad. Sci. USA. 100, 5968–5973. https://doi.org/10.1073/pnas.1037651100
Chevigny N., Schatz-Daas D., Lotfi F., Gualberto J.M. (2020) DNA repair and the stability of the plant mitochondrial genome. Int. J. Mol. Sci. 21, 328. https://doi.org/10.3390/ijms21010328
Woloszynska M., Trojanowski T. (2009) Counting mtDNA molecules in Phaseolus vulgaris: sublimons are constantly produced by recombination via short repeats and undergo rigorous selection during substoichiometric shifting. Plant Mol. Biol. 70, 511–521. https://doi.org/10.1007/s11103-009-9488-8
Newton K.J., Gabay-Laughnan S., De Paepe R. (2004) Mitochondrial mutations in plants. In: Plant Mitochondria: From Genome to Function. Advances in Photosynthesis and Respiration. 17, 121–141. https://doi.org/10.1007/978-1-4020-2400-9_7
Andersson S. (1999) Quantitative genetics of leaf morphology in Crepis tectorum ssp. pumila (Asteraceae). J. Heredity. 90, 556–561. https://doi.org/10.1093/jhered/90.5.556
Kajander O.A., Rovio A.T., Majamaa K., Poulton J., Spelbrink J.N., Holt I.J., Karhunen P.J., Jacobs H.T. (2000) Human mtDNA sublimons resemble rearranged mitochondrial genomes found in pathological states. Hum. Mol. Genet. 22, 2821–2835. https://doi.org/10.1093/hmg/9.19.2821
Knorre D.A., Popadin K.Y., Sokolov S.S., Severin F.F. (2013) Roles of mitochondrial dynamics under stressful and normal conditions in yeast cells. Oxid. Med. Cell Longev. 2013, 139491. https://doi.org/10.1155/2013/139491
Christensen A.C. (2013) Plant mitochondrial genome evolution can be explained by DNA repair mechanisms. Genome Biol. Evol. 5, 1079–1086. https://doi.org/10.1093/gbe/evt069
Boesch P., Weber-Lotfi F., Ibrahim N., Tarasenko V., Cosset A., Paulus F., Lightowlers R.N., Dietrich A. (2011) DNA repair in organelles: pathways, organization, regulation, relevance in disease and aging. Biochim. Biophys. Acta. 1813, 186–200. https://doi.org/10.1016/j.bbamcr.2010.10.002
Cappadocia L., Maréchal A., Parent J.S, Lepage E., Sygusch J., Brisson N. (2010) Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell. 22, 1849–1867. https://doi.org/10.1105/tpc.109.071399
Zaegel V., Guermann B., Le Ret M., Andres C., Meyer D., Erhardt M., Canaday J., Gualberto J.M., Imbault P. (2006) The plant-specific ssDNA binding protein OSB1 is involved in the stoichiometric transmission of mitochondrial DNA in Arabidopsis. Plant Cell. 18, 3548–3563. https://doi.org/10.1105/tpc.106.042028
Shedge V., Arrieta-Montiel M., Christensen A.C., Mackenzie S.A. (2007) Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell. 19, 1251–1264. https://doi.org/10.1105/tpc.106.048355
Wu Z., Waneka G., Broz A.K., King C.R., Sloan D.B. (2020) MSH1 is required for maintenance of the low mutation rates in plant mitochondrial and plastid genomes. Proc. Natl. Acad. Sci. USA. 117, 16448–16455. https://doi.org/10.1073/pnas.2001998117
Ladoukakis E.D., Zouros E. (2001) Direct evidence for homologous recombination in mussel (Mytilus galloprovincialis) mitochondrial DNA. Mol. Biol. Evol. 18, 1168‒1175. https://doi.org/10.1093/oxfordjournals.molbev.a003904
Ramsey A.J., Mandel J.R. (2019) When one genome is not enough: organellar heteroplasmy in plants. Ann. Plant Rev. Online. 2(2). https://doi.org/10.1002/9781119312994.apr0616
Aksyonova E., Sinyavskaya M., Danilenko N., Per-shina L., Nakamura C., Davydenko O. (2005) Heteroplasmy and paternally oriented shift of the organellar DNA composition in barley–wheat hybrids during backcrosses with wheat parents. Genome. 48, 761–769. https://doi.org/10.1139/g05-049
Khrapko K., Coller H.A., André P.C., Li. X.C., Hanekamp J.S., Thilly W.G. (1997) Mitochondrial mutational spectra in human cells and tissues. Proc. Natl. Acad. Sci. USA. 94, 13798–13803. https://doi.org/10.1073/pnas.94.25.13798
Garcia-Diaz A., Oya R., Sanchez A., Luque F. (2003) Effect of prolonged vegetative reproduction of olive tree cultivars (Olea europaea L.) in mitochondrial homoplasmy and heteroplasmy. Genome. 46, 377–381. https://doi.org/10.1139/g03-017
Li X.Q., Chetrit P., Mathieu C., Vedel F., De Paepe R., Remy R., Ambard-Bretteville F. (1988) Regeneration of cytoplasmic male-sterile protoclones of Nicotiana sylvestris with mitochondrial variations. Curr. Genet. 13, 261–266. https://doi.org/10.1007/BF00387773
Pla M., Mathieu C., De Paepe R., Chetrit P., Vedel F. (1995) Deletion of the last two exons of the mitochondrial nad7 gene results in lack of the NAD7 polypeptide in a Nicotiana sylvestris CMS mutant. Mol. Gen. Genet. 248, 79–88. https://doi.org/10.1007/BF02456616
Guo Y., Li C., Sheng Q., Winther J.F., Cai Q., Boice J.D., Shyr Y. (2013) Very low-level heteroplasmy mtDNA variations are inherited in humans. J. Genet. Genomics. 40, 607–615. https://doi.org/10.1016/j.jgg.2013.10.003
Stewart J.B. Chinnery P.F. (2015) The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat. Rev. Genet. 16, 530–542. https://doi.org/10.1038/nrg3966
Zhang Y., Wang C., Jin Y., Yang Q., Meng Q., Liu Q., Dai Y., Cai L., Liu Z., Liu K., Sun H. (2018) Activa-ting the PGC-1 α/TERT pathway by Catalpol ameliorates atherosclerosis via modulating ROS production, DNA damage, and telomere function: implications on mitochondria and telomere link. Oxidat. Med. Cell. Longevity. 2018, 2876350. https://doi.org/10.1155/2018/2876350
Sekiguchi K., Imaizumi K., Matsuda H., Mizuno N., Yoshida K., Senju H., Sato H., Kasai K. (2003) MtDNA sequence analysis using capillary electrophoresis and its application to the analysis of mtDNA in hair. Japanese Journal of Science and Technology for Identification. 7, 123–130. https://doi.org/10.3408/jasti.7.123
Wilton P.R., Zaidi A., Makova K., Nielsen R. (2018) A population phylogenetic view of mitochondrial heteroplasmy. Genetics. 208, 1261–1274. https://doi.org/10.1534/genetics.118.300711
Jenuth J.P., Peterson A.C., Fu K., Shoubridge E.A. (1996) Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat. Genet. 14, 146–151. https://doi.org/10.1038/ng1096-146
Chinnery P.F. (2002) Modulating heteroplasmy. Trends Genet. 18, 173–176. https://doi.org/10.1016/s0168-9525(01)02636-1
Johnston I.G. (2019b) Varied mechanisms and models for the varying mitochondrial bottleneck. Front. Cell Dev. Biol. 7, 294. https://doi.org/10.3389/fcell.2019.00294
Johnston I.G., Jones N.S. (2016) Evolution of cell-to-cell variability in stochastic, controlled, heteroplasmic mtDNA populations. Am. J. Hum. Genet. 99, 1150–1162. https://doi.org/10.1016/j.ajhg.2016.09.016
Galtier N. (2011) The intriguing evolutionary dyna-mics of plant mitochondrial DNA. BMC Biol. 9, 61. https://doi.org/10.1186/1741-7007-9-61
Kuroiwa H., Kuroiwa T. (1992) Giant mitochondria in the mature egg cell of Pelargonium zonale. Protoplasma. 168, 184–188.https://doi.org/1007/BF01666264
Kanazawa A., Tsutsumi N., Hirai A. (1994) Reversible changes in the composition of the population of mtDNAs during dedifferentiation and regeneration in tobacco. Genetics. 138, 865–870. https://doi.org/10.1093/genetics/138.3.865
Arrieta-Montiel M., Lyznik A., Woloszynska M., Janska H., Tohme J., Mackenzie S. (2001) Tracing evolutionary and developmental implications of mitochondrial stoichiometric shifting in the common bean. Genetics. 158, 851–864. https://doi.org/10.1093/genetics/158.2.851
Albert B., Lelandais C., Pla M., Leuret C., Vitart V., Mathieu C., Sihachakr D., Godelle B., de Paepe R. (2003) Amplification of Nicotiana sylvestris mitochondrial subgenomes is under nuclear control and is associated with phenotypic changes. Genetica. 117, 17–25. https://doi.org/10.1023/a:1022356330794
Suzuki T., Kawano S., Sakai A., Hirai A., Kuroiwa T. (1996) Variability of mitochondrial subgenomic molecules in the meristematic cells of higher plants. Genes Genet. Syst. 71, 329–333. https://doi.org/10.1266/ggs.71.329
Velappan Y., Signorelli S., Considine M.J. (2017) Cell cycle arrest in plants: what distinguishes quiescence, dormancy and differentiated G1? Ann. Bot. 120, 495–509. https://doi.org/10.1093/aob/mcx082
Chinnery P.F., Turnbull D.M. (1999) Mitochondrial DNA and disease. Lancet. 354, S17–S21. https://doi.org/10.1016/s0140-6736(99)90244-1
Taylor R.W. (2005) Gene therapy for the treatment of mitochondrial DNA disorder. Expert. Opin. Biol. Ther. 5, 183–194. https://doi.org/10.1517/14712598.5.2.183
Trifunovic A., Larsso N.-G. (2008) Mitochondrial dysfunction as a cause of ageing. J. Intern. Med. 263, 167–178. https://doi.org/10.1111/j.1365-2796.2007.01905.x
Wallace D.C. (2010) Mitochondrial DNA mutations in disease and aging. Environ. Mol. Mutagen. 51, 440–450. https://doi.org/10.1002/em.20586
Li M., Schönberg A., Schaefer M., Schroeder R., Nasidze I., Stoneking M. (2010) Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am. J. Hum. Genet. 87, 237–249. https://doi.org/10.1016/j.ajhg.2010.07.014
Li M., Rothwell R., Vermaat M., Wachsmuth M., Schröder R., Laros J.F.J., van Oven M., de Bakker P.I.W., Bovenberg J.A., van Duijn C.M., van Ommen G.-J., Slagboom P.E., Swertz M.A., Wijmenga C., Kayser M., Boomsma D.I., Zöllner S., de Knijff P., Stoneking M. (2016) Transmission of human mtDNA heteroplasmy in the genome of the Netherlands families: support for a variable-size bottleneck. Genome Res. 26, 417–426. https://doi.org/10.1101/gr.203216.115
Rebolledo-Jaramillo B., Su M.S.-W., Stoler N., McElhoe J.A., Dickins B., Blankenberg D., Korneliussen T.S., Chiaromonte F., Nielsen R., Holland M.M., Paul I.M., Nekrutenko A., Makova K.D. (2014) Maternal age effect and severe germ-line bottleneck in the inheritance of human mitochondrial DNA. Proc. Natl. Acad. Sci. USA. 111, 15474–15479. https://doi.org/10.1073/pnas.1409328111
Gu J., Miles D., Newton K.J. (1993) Analysis of leaf sectors in the NCS6 mitochondrial mutant of maize. Plant Cell. 5, 963–971. https://doi.org/10.1105/tpc.5.8.963
Sakamoto W., Kondo H., Murata M., Motoyoshi F. (1996) Altered mitochondrial gene expression in a maternal distorted leaf mutant of Arabidopsis induced by chloroplast mutator. Plant Cell. 8, 1377–1390. https://doi.org/10.1105/tpc.8.8.1377
Bellaoui M., Martin-Canadell A., Pelletier G., Budar F. (1998) Lowcopy-number molecules are produced by recombination, actively maintained and can be ampliWed in the mitochondrial genome of Brassicaceae: relationship to reversion of the male sterile phenotype in some cybrids. Mol. Gen. Genet. 257, 177–185. https://doi.org/10.1007/s004380050637
Vitart V., De Paepe R., Mathieu C., Chetrit P., Vedel F. (1992) Amplification of substoichiometric recombinant mitochondrial DNA sequences in a nuclear, male sterile mutant regenerated from protoplast culture in Nicotiana sylvestris. Mol. Gen. Genet. 233, 193–200. https://doi.org/10.1007/BF00587579
Hartmann C., Recipon H., Jubier M.F., Valon C., Delcher-Besin E., Henry Y., De Buyser J., Lejeune B., Rode A. (1994) Mitochondrial DNA variability detected in a single wheat regenerant involves a rare recombination event across a short repeat. Curr. Genet. 25, 456–464. https://doi.org/10.1007/BF00351786
Taylor D.R., Olson M.S., McCauley D.E. (2001) A quantitative genetic analysis of nuclear-cytoplasmic male sterility in structured populations of Silene vulgaris. Genetics. 158, 833–841. https://doi.org/10.1093/genetics/158.2.833
Welch M.E., Darnell M.Z., McCauley D.E. (2006) Variable populations within variable populations: quantifying mitochondrial heteroplasmy in natural populations of the gynodioecious plant Silene vulgaris. Genetics. 174, 829–837. https://doi.org/10.1534/genetics.106.059246
Frey J.E., Frey B., Forcioli D. (2005) Quantitative assessment of heteroplasmy levels in Senecio vulgaris chloroplast DNA. Genetica. 123, 255–261. https://doi.org/10.1007/s10709-004-3711-y
Pearl S.A., Welch M.E., McCauley D.E. (2009) Mitochondrial heteroplasmy and paternal leakage in natural populations of Silene vulgaris, a gynodioecious plant. Mol. Biol. Evol. 26, 537–545. https://doi.org/10.1093/molbev/msn273
Mandel J.R., McCauley D.E. (2015) Pervasive mitochondrial sequence heteroplasmy in natural populations of wild carrot, Daucus carota spp. carota L. PLoS One. 10, e0136303. https://doi.org/10.1371/journal.pone.0136303
Mishra P., Chan D.C. (2016) Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 212, 379–387. https://doi.org/10.1083/jcb.201511036
Jakobs S., Martini N., Schauss A.C., Egner A., Westermann B., Hell S.W. (2003) Spatial and temporal dynamics of budding yeast mitochondria lacking the division component Fis1p. J. Cell Sci. 116, 2005–2014. https://doi.org/10.1242/jcs.00423
Tondera D., Grandemange S., Jourdain A., Karbowski M., Mattenberger Y., Herzig S., Da Cruz S., Clerc P., Raschke I., Merkwirth C., Ehses S., Krause F., Chan D.C., Alexander C., Bauer C., Youle R., Langer T., Martinou J.-C. (2009) SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 28, 1589–1600. https://doi.org/10.1038/emboj.2009.89
Mitra K., Wunder C., Roysam B., Lin G., Lippincott-Schwartz J. (2009) A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc. Natl. Acad. Sci. USA. 106, 11960–11965. https://doi.org/10.1073/pnas.0904875106
Yao C.-H., Wang R., Wang Y., Kung C.-P., Weber J.D., Patti G.J. (2019) Mitochondrial fusion supports increased oxidative phosphorylation during cell prolife-ration. Elife. 8, e41351. https://doi.org/10.7554/eLife.41351
Ramonell K.M., Kuang A., Porterfield D.M., Crispi M.L., Xiao Y., McClure G., Musgrave M.E. (2001) Influence of atmospheric oxygen on leaf structure and starch deposition in Arabidopsis thaliana. Plant Cell Environ. 24, 419–428. https://doi.org/10.1046/j.1365-3040.2001.00691.x
May A.I., Devenish R.J., Prescott M. (2012) The many faces of mitochondrial autophagy: making sense of contrasting observations in recent research. Int. J. Cell Biol. 2012, 431684. https://doi.org/10.1155/2012/431684
Popkov V.A., Plotnikov E.Y., Lyamzaev K.G., Silachev D.N., Zorova L.D., Pevzner I.B., Jankauskas S.S., Zorov S.D., Babenko V.A., Zorov D.B. (2015) Mitodiversity. Biochemistry (Mosc.). 80, 532‒541. https://doi.org/10.1134/S000629791505003X
Li F., Chung T., Vierstra R.D. (2014) Autophagy-related11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell. 26, 788–807. https://doi.org/10.1105/tpc.113.120014
Minibayeva F., Dmitrieva S., Ponomareva A., Ryabovol V. (2012) Oxidative stress-induced autophagy in plants: the role of mitochondria. Plant Physiol. Biochem. 59, 11–19. https://doi.org/10.1016/j.plaphy.2012.02.013
Wikstrom J.D., Katzman S.M., Mohamed H., Twig G., Graf S.A., Heart E., Molina A.J.A., Corkey B.E., Moitoso de Vargas L., Danial N.N., Collins S., Shirihai O.S. (2007) beta-Cell mitochondria exhibit membrane potential heterogeneity that can be altered by stimulatory or toxic fuel levels. Diabetes. 56, 2569–2578. https://doi.org/10.2337/db06-0757
Distelmaier F., Koopman W.J., Testa E.R., de Jong A.S., Swarts H.G., Mayatepe E., Smeitink J.A.M., Willems P.H.G. (2008) Life cell quantification of mitochondrial membrane potential at the single organelle level. Cytometry A. 73, 29–38. https://doi.org/10.1002/cyto.a.20503
Law S.R., Narsai R., Taylor N.L., Delannoy E., Carrie C., Giraud E., Millar A.H., Small I., Whelan J. (2012) Nucleotide and RNA metabolism prime translational initiation in the earliest events of mitochondrial biogenesis during Arabidopsis germination. Plant Physiol. 158, 1610–1627. https://doi.org/10.1104/pp.111.192351
Konstantinov Y.M., Dietrich A., Weber-Lotfi F., Ibrahim N., Klimenko E.S., Tarasenko V.I., Bolotova T.A., Koulintchenko M.V. (2016) DNA import into mitochondria. Biochemistry (Mosc.). 81, 1044–1056. https://doi.org/10.1134/S0006297916100035
Armstrong A.F., Logan D.C., Tobin A.K., O’Toole P., Atkin O.K. (2006) Heterogeneity of plant mitochondrial responses underpinning respiratory acclimation to the cold in Arabidopsis thaliana leaves. Plant Cell Environ. 29, 940–949. https://doi.org/10.1111/j.1365-3040.2005.01475.x
Tarasenko T.A., Klimenko E.S., Tarasenko V.I., Koulintchenko M.V., Dietrich A., Weber-Lotfi F., Konstantinov Y.M. (2021) Plant mitochondria import DNA via alternative membrane complexes involving various VDAC isoforms. Mitochondrion. 60, 43–58. https://doi.org/10.1016/j.mito.2021.07.006
Wade M.J., McCauley D.E. (2005) Paternal leakage sustains the cytoplasmic polymorphism underlying gynodioecy but remains invasible by nuclear restorers. Am. Nat. 166, 592‒602. https://doi.org/10.1086/491660
McCauley D.E. (2013) Paternal leakage, heteroplasmy, and the evolution of plant mitochondrial genomes. New Phytol. 200, 966‒977. https://doi.org/10.1111/nph.12431
Christie J.R., Schaerf T.M., Beekman M. (2015) Selection against heteroplasmy explains the evolution of uniparental inheritance of mitochondria. PLoS Genet. 11, e1005112. https://doi.org/10.1371/journal.pgen.1005112
Дополнительные материалы отсутствуют.
Инструменты
Молекулярная биология


