Российский физиологический журнал им. И.М. Сеченова, 2023, T. 109, № 11, стр. 1522-1546
Молекулярно-генетические механизмы регуляции циркадных ритмов и их роль в психопатологиях
К. В. Смирнова 1, 2, *, Н. Д. Чижова 1, 2, Е. В. Герасимова 3, А. В. Калуев 1, 2, 3, 4, 5, 6, Т. Г. Амстиславская 1, 2, **
1 Научно-исследовательский институт нейронаук и медицины
Новосибирск, Россия
2 Новосибирский государственный университет
Новосибирск, Россия
3 Научно-технологический университет “Сириус”
Сочи, Россия
4 Санкт-Петербургский государственный университет
Санкт-Петербург, Россия
5 Уральский федеральный университет
Екатеринбург, Россия
6 Национальный медицинский исследовательский центр им. В.А. Алмазова МЗ РФ
Санкт-Петербург, Россия
* E-mail: vedelina@mail.ru
** E-mail: amstislavskayatg@neuronm.ru
Поступила в редакцию 07.12.2022
После доработки 12.01.2023
Принята к публикации 14.01.2023
- EDN: GNPRKI
- DOI: 10.31857/S0869813923110109
Аннотация
Циркадные (циркадианные) ритмы представляют собой циклические колебания интенсивности биологических процессов, связанные со сменой дня и ночи, к которым в ходе эволюции приспособились многие живые организмы. Нарушения циркадных ритмов провоцируются как факторами среды (смена часового пояса или продолжительности дня и ночи), так и поломками во внутренней регуляции циклов (мутации ключевых “часовых” генов). Эти изменения могут приводить к патогенезу различных заболеваний, в том числе психопатологий. Поскольку механизмы, лежащие в основе циркадной регуляции, достаточно консервативны, для большего понимания этих процессов и их связи с психопатологиями активно используют экспериментальные модели in vivo. В настоящем обзоре рассмотрены вопросы регуляции циркадных ритмов, а также их межтаксонные сходства и различия у млекопитающих и костных рыб (на примере широко используемых в биомедицине рыб зебраданио, Danio rerio). В работе обсуждаются современные представления о молекулярно-генетических механизмах, лежащих в основе регуляции циркадных ритмов, и их взаимосвязь с патогенезом психических расстройств у человека и модельных организмов.
ВЕДЕНИЕ
Циркадные (циркадианные) ритмы представляют собой широкий спектр поведенческих, гормональных и биохимических процессов, возникающих под влиянием изменения длительности светового дня [1]. В головном мозге позвоночных существует главный водитель ритма – парное супрахиазматическое ядро гипоталамуса (SCN), которое контролирует биоритмы всех систем органов, тканей и клеток организма, определяя время основных физиологических событий (пищеварения, сна и бодрствования, моторной активности и др.) [2]. На молекулярном уровне регуляция циркадных ритмов представляет собой петлю отрицательной обратной связи. Ключевыми ее участниками выступают транскрипционные факторы CLOCK и BMAL1 – позитивные регуляторы экспрессии генов period (Per), cryptochrome (Cry) и reverseerythoblastoma (Rev-Erba α/β), белковые продукты которых, в свою очередь, являются репрессорами транскрипции Clock и Bmal1 (Arntl1) [1].
Люди с определенными полиморфизмами в генах циркадной регуляции имеют предрасположенность к психическим расстройствам, которые также могут возникать при нарушении циркадных ритмов вследствие частых перелетов или работы в ночную смену [3, 4]. Высокая степень эволюционной консервативности процессов циркадной регуляции у разных видов позвоночных позволила создать валидные экспериментальные модели психопатологий с генетическим или средовым нарушением циркадных ритмов [1]. И хотя в качестве классического объекта для таких исследований используют грызунов, все чаще для изучения функций мозга начинают использовать пресноводных костных рыб зебраданио (zebrafish, Danio rerio) [5, 6].
В настоящем обзоре рассмотрены современные данные о связи циркадных ритмов и психопатологий, в качестве факторов патогенеза которых выступают полиморфизмы циркадных генов и средовые нарушения суточных ритмов. Обсуждаются также экспериментальные (животные) модели психопатологий, связанные с поломками циркадных ритмов позвоночных. В работе показана общность молекулярно-генетических механизмов, лежащих в основе регуляции циркадных ритмов у эволюционно отстоящих друг от друга классов (млекопитающих и костных рыб), что открывает возможности для развития направления трансляционной биологии и использования полученных данных в практической медицине.
РОЛЬ СВЕТА В ЦИРКАДНОЙ АКТИВНОСТИ СИСТЕМ ОРГАНИЗМА
Главный внешний фактор, влияющий на циркадные ритмы – это изменение окружающего освещения, которое оказывает влияние на SCN. У млекопитающих это ядро получает световые сигналы от специальных клеток сетчатки – меланопсин-экспрессирующих фоточувствительных ганглиозных клеток (ipRGC), дающих проекции в мозг, тем самым опосредуя широкий спектр зрительных функций [7]. Меланопсин, связанный с G-белком, возбуждается светом, в результате чего мембраны ipRGC деполяризуются, модулируя физиологические и поведенческие процессы [8]. IpRGC характерны для многих позвоночных, в т.ч. зебраданио. И хотя их устройство и принцип работы похожи у представителей разных таксонов, имеются некоторые различия в генах, кодирующих меланопсин. В частности у млекопитающих он кодируется всего одним геном из семейства Opn4m, тогда как у рыб (зебраданио) – тремя генами из семейства opn4m и двумя генами из семейства opn4x [9].
Важно отметить, что меланопсин млекопитающих имеет максимум абсорбции при длине волны 467 нм (синий свет), после чего он преобразуется в продукт с максимумом абсорбции при 476 нм (синий свет). Существует также особое состояние меланопсина – экстрамеланопсин, который образуется при длинноволновом свете более 575 нм (от желтого до красного света) и может возвращаться в обычное состояние после воздействия синего света [10]. Это важно в установке циркадного ритма млекопитающих, поскольку SCN в большей степени отслеживает изменения спектрального состава света между синим и желтым, нежели интенсивность освещения [11]. В целом широкая спектральная чувствительность ipRGC достигает максимума при 460 нм, что аналогично распределению длин волн в сумерках [8]. В отличие от млекопитающих, все пять генов меланопсинов зебраданио кодируют функциональный фотопигмент с пиковой спектральной чувствительностью в диапазоне от 470 до 484 нм, при этом гены opn4m-1 и opn4m-3 демонстрируют бистабильность, как у беспозвоночных, где хромофор сетчатки меняет цис- и меры, а продукты активности opn4m-2, opn4x-1 и opn4x-2 являются моностабильными и функционируют скорее как классические фотопигменты позвоночных [9]. Периферические клетки зебраданио также экспрессируют большое количество различных опсинов и реагируют на разные длины волн, способные запускать экспрессию циркадных генов, включая ультрафиолетовое и инфракрасное излучение [12].
Характерной чертой млекопитающих является иерархическое устройство циркадной системы, где SCN играет роль главного водителя ритма (рис. 1). Полученные от ipRGC сигналы моносинаптически передаются по ретиногипоталамическому тракту в SCN. Ядро представляет собой гетерогенную структуру, чьи нейроны анатомически и функционально разделены на две области: венотромедиальную сердцевину и дорзолатеральную оболочку, основными нейропептидами которых являются вазоактивный интестинальный полипептид (VIP) и аргинин-вазопрессин (AVP) соответственно [13]. Нейроны SCN могут также содержать тормозный нейромедиатор – гамма-аминомасляную кислоту (ГАМК) [14, 15]. VIP-содержащие нейроны после получения информации от сетчатки дают проекции в AVP-содержащие клетки SCN. Посредством рецепторов VPAC2 они удерживают синхронизацию клеточной активности ядра в целом [13, 16]. AVP-содержащие нейроны координируют другие структуры мозга и органы через нейроэндокринную гипоталамо-гипофизарно-надпочечниковую ось (HPA), согласовывая поведенческие и физиологические ритмы [17, 18]. При этом ipRGC сетчатки могут давать проекции не только в SCN, поскольку у мышей было показано, что сигналы от ipRGC могут поступать в дорзальное перихабенулярное ядро, а оттуда в прилежащее ядро (NAc), тем самым участвуя в регуляции поведения [19].
Рис. 1.
Иерархическое устройство циркадной системы млекопитающих и запуск циркадных колебаний светом через ipRGC. Белые стрелки – воздействие света, серые – проекции нейронов, черные – биохимические пути внутри SCN.
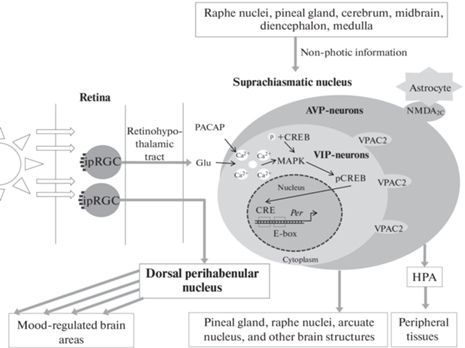
SCN имеет связи со множеством зон мозга, наиболее значимой из которых является шишковидная железа, вырабатывающая мелатонин. Нарушение этих связей в результате нейродегенеративных процессов приводит к нарушению циркадных ритмов [15]. Помимо этого, ядро отсылает проекции в другие зоны гипоталамуса, серотонинергические ядра шва и дугообразное ядро [20–22]. SCN также получает дополнительные входы от ядер шва (через шовно-гипоталамический тракт), от шишковидной железы и от многочисленных областей конечного, промежуточного, среднего и продолговатого мозга, обеспечивая взаимосвязь циркадных ритмов и поведения [23].
Важно отметить, что активность SCN регулируется не только нейронами, но и астроцитами. Так, например, нейроны SCN проявляют активность в течение светового дня, а астроциты работают ночью и могут подавлять активность нейронов дорзальной части SCN через регуляцию комплекса пресинаптических глутаматергических N-метил-D-аспартатных рецепторов NMDA2C [24]. Поскольку астроциты, как и глия в целом, активно вовлечены в развитие психопатологий, такое взаимодействие с SCN порождает дополнительную связь между циркадными ритмами и поведенческими нарушениями [25].
В отличие от млекопитающих, у костных рыб светочувствительные клетки расположены по всему телу, а сама циркадная система децентрализована. Внутри мозга костных рыб обнаружены глубинно расположенные фоторецепторы, например, в таламусе обнаружена экспрессия некоторых генов опсинов, в том числе va-opsin, val-opsin, tmt-opsin и opn4 [26]. В других областях также обнаружены фоторецепторы [27]. Фоточувствительные нейроны преоптической области личинок зебраданио позволяют им улавливать свет даже в отсутствие глаз, они вовлечены в реакцию поиска света и участвуют в выборе стратегии поведения при этом поиске [28, 29]. Активность глубинно расположенных фоторецепторов костных рыб может изменяться в течение дня [30]. Тем не менее, функции этих рецепторов в циркадной регуляции остаются малоизученными, что представляет собой интересную область для кросс-таксонных нейробиологичесих исследований.
Основные зоны мозга, вовлеченные в циркадную регуляцию у рыб, не являются необходимыми для функционирования ритмов периферических тканей [31]. У рыб уже в первые 16 ч после оплодотворения световой режим влияет на ритм экспрессии некоторых мРНК, тогда как шишковидная железа формируется на 3–4 дня позже [32]. Эта особенность существенно отличает костных рыб от млекопитающих, у которых синхронизация ритмов происходит во время эмбрионального и раннего неонатального развития за счет циркадной системы матери, которая координирует синхронизацию циркадных часов в SCN плода [33]. Несмотря на децентрализованность циркадной системы у костных рыб, у них есть области мозга, аналогичные SCN млекопитающих, которые также могут получать информацию как от сетчатки, так и от других структур мозга, в частности шишковидной железы. Далее информация передается в передний и средний мозг, а также в переднее прегломерулярное ядро. При этом шишковидная железа рыб имеет свой собственный осциллятор и может функционировать без SCN, совершая ритмическую выработку мелатонина и определяя ритмические паттерны поведения. Поскольку большинство мишеней шишковидной железы зебраданио, по-видимому, являются премоторными и прецеребеллярными центрами, то ее проекции участвуют в световой и циркадной модуляции этих центров [27, 34, 35]. Таким образом, существует базовое сходство в структурах мозга и их функциональных связях между зебраданио и млекопитающими, позволяя использовать их как экспериментальные модели для исследования циркадных процессов.
МОЛЕКУЛЯРНЫЕ МЕХАНИЗМЫ ЦИРКАДНЫХ ЧАСОВ МЛЕКОПИТАЮЩИХ И КОСТНЫХ РЫБ
Биохимические компоненты циркадных часов различаются в зависимости от таксона, но основаны на ритмичном изменении концентрации факторов по принципу отрицательной обратной связи (рис. 2.). Это колебание управляет суточными паттернами экспрессии генов, которые регулируют метаболические ритмы клеток организма [1, 36]. Циркадные часы большинства позвоночных имеют несколько основных транскрипционных факторов, участвующих в суточных осцилляциях: CLOCK и BMAL1 – активаторы транскрипции циркадных генов и семейства белков PER и CRY – репрессоры транскрипции [1].
Рис. 2.
Молекулярные механизмы циркадных часов млекопитающих. Толстые сплошные и прерывистые стрелки иллюстрируют основные молекулярные пути, тонкие – дополнительные. Прерывистые стрелки – выход мРНК из ядра и трансляция. Крест – блокирование транскрипции. Отрезки с круглым окончанием обозначают содействие процессу определенными белками. Прямоугольные участки на ДНК – промоторы.
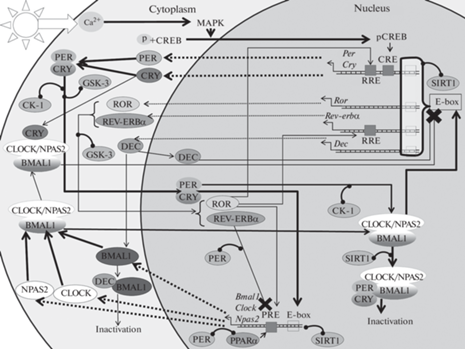
У млекопитающих клеточные осцилляции запускаются светом через ретиногипоталамический тракт с помощью глутамата, который увеличивает внутриклеточную концентрацию Ca2+ в нейронах SCN, тем самым активируя MAPK-сигнальный каскад. Предположительно, в этом процессе принимает участие пептид, активирующий аденилатциклазу гипофиза (PACAP) [37–39]. Это приводит к фосфорилированию транскрипционного фактора CREB и его транслокации в ядро, где он связывается с элементом CRE на промоторе ряда генов, в том числе Per [37, 38]. В течение дня в цитоплазме накапливаются белки PER, они образуют комплекс с CRY и транслоцируются в ядро, где активируют транскрипцию Bmal1 и Clock [40, 41]. Важно отметить, что PER нуждается в CRY для своего ядерного транспорта, тогда как CRY могут проходить в ядро самостоятельно [42]. Экспрессия Bmal1 регулируется не только за счет PER2, но и благодаря ROR-элементу (RRE) на промоторе этого гена. Так, REV-ERBα играет ключевую роль в регуляции колебаний экспрессии Bmal1, ингибируя его транскрипцию, а белок RORα активирует ее, связываясь с RRE [43]. Аналогично регулируется экспрессия других часовых генов, Npas2 и Clock [44].
PER2 способен напрямую взаимодействовать с ядерными рецепторами REV-ERBα или PPARα на RRE промоторах Bmal1, действуя как корепрессор или коактиватор соответственно [41]. Гетеродимер белков CLOCK (или его паралог NPAS2) и BMAL1 способны связываться с E-бокс-регуляторным элементом на промоторах различных генов, в том числе Rev-erbα, Ror, Per, Dec и Cry, активируя их транскрипцию. Накопление белков PER и CRY, их последующая димеризация, транспорт в ядро и присоединение к комплексу CLOCK:BMAL1 нарушает связь последнего с промоторами генов и останавливает транскрипцию [45, 46]. CRY способны связываться с CLOCK:BMAL1 в цитоплазме и блокировать активацию генов мишеней CLOCK:BMAL1, не нарушая при этом связи комплекса с промоторами [46, 47]. Dес1 и Dес2 представляют собой гены, кодирующие факторы транскрипции bHLH (Basic-helix-loop-helix), которые ингибируют Per и свою собственную транскрипцию за счет связывания с белком BMAL1 и/или конкуренции с CLOCK:BMAL1 за последовательности E-бокса на своих собственных промоторах. Считается, что эти молекулярные петли обеспечивают стабильность молекулярных часов и их точную настройку [36]. Так, DEC1,2 совместно с белком CHRONO контролирует длину циркадного периода в SCN новорожденных мышей. CHRONO участвует в объединении клеточных колебаний с разными периодами, а DEC1,2 удлиняет период раннего неонатального периода за счет интеграции клеточных ритмов [48].
Циркадный осциллятор зебраданио во многом схож с таковым у млекопитающих. Главным различием в его молекулярных механизмах является то, что костные рыбы в ходе эволюции претерпели дупликацию генома, что оказало влияние на циркадную генетику. Так, например, зебраданио имеют по 2 копии генов arntl1 (arntl1a и arntl1b, или bmal1a/b) и clock (clock и clock-3), однако эти копии имеют сходную циркадную фазу [49]. Тем не менее у костных рыб обнаружена диверсифицированная регуляция экспрессии среди многих копий канонических часовых генов, что проявляется в тканеспецифических паттернах экспрессии в ответах на внутренние и внешние стимулы. В итоге, эти различия, вероятно, отражают неравномерность влияния суточных факторов на метаболизм и клеточные процессы в центральных и периферических тканях рыб [50]. Если кратко представить молекулярный осциллятор зебраданио, то процесс похож на таковой у млекопитающих, еще раз подтверждая консервативность процессов циркадной регуляции. В частности, гетеродимер CLOCK:BMAL1 активирует транскрипцию генов per2 и cry1a, их белковые продукты димеризуются, соединяются с комплексом CLOCK:BMAL1, что подавляет активацию транскрипции этих генов [32]. Белок Per2 играет двойную роль в регуляции циркадных часов зебраданио. Он репрессирует экспрессию гена шишковидной железы арилалкиламин N-ацетилтрансферазы 2 (aanat2) через E-box и снижает/усиливает экспрессию arntl1b посредством связывания с Rev-erb/Rorα соответственно.
Более того, Per2 рыб зебраданио, по-видимому, также выполняет тканеспецифические регуляторные роли во многих периферических органах и имеет решающее значение для поддержания циркадной регуляции в сердце и печени [51]. Для зебраданио также важна передача по сигнальному пути TGF-β для нормальной функции циркадных часов, поскольку TGF-β-сигналинг оказывает влияние на фазу экспрессии per1b [52]. У млекопитающих данный путь участвует в сопряжении периферических тканей, опосредуя паракринную настройку фазы молекулярных часов и регулируя транскрипции основных часовых генов [53]. Сходство процессов циркадной регуляции рыб и млекопитающих в очередной раз указывает на эволюционную древность этих механизмов.
Следует отметить, что фазы экспрессии циркадных генов могут различаться у дневных и ночных животных – как, например, происходит у мышей (Mus musculus, ведущих преимущественно ночной образ жизни) и бабуинов (Papio anubis, представителей дневных животных), пики экспрессии основных часовых генов которых различаются по фазе примерно на 12 ч. Гены Bmal1 и Per1 у приматов показывают пики экспрессии вечером и утром соответственно, тогда как у мышей эти пики демонстрируют обратную картину в большинстве тканей организма. Интересно, что Cry1 отличается от других генов: средняя фаза его экспрессии у бабуинов приходится на конец дня, а у мышей задерживается на 7 ч до пика после полуночи. Важно отметить, что в SCN в отличие от других тканей фаза экспрессии компонентов циркадных часов мышей не отличается от таковой у бабуинов. Так, фаза пика экспрессии Bmal1 одинаково приходится на ранний вечер и у бабуинов, и у мышей, а фазы экспрессии Per1 и Cry2 проходят через 6 и 12 ч соответственно [54]. Это показывает, что в периферических тканях млекопитающих происходит сложная настройка суточных ритмов, как упоминалось выше, и этот фактор следует учитывать при планировании экспериментов.
ПОСТТРАНСКРИПЦИОННЫЕ И ПОСТТРАНСЛЯЦИОННЫЕ МОДИФИКАЦИИ В РЕГУЛЯЦИИ СУТОЧНЫХ РИТМОВ
Большую роль в циркадной регуляции играют посттранскрипционные и посттрансляционные модификации. Например, менее 30% мРНК циркадных генов регулируются транскрипцией de novo, что предполагает существование механизмов посттранскрипционной циркадной регуляции [55]. Доказательством этому служит то, что некоторые мРНК-редактирующие ферменты имеют ритмическую экспрессию [56]. Так, нарушение образования кэп-структуры на 5'-конце молекулы мРНК у мышей приводит к пролонгированию циркадного периода часовых генов [57]. Стабильность циркадных мРНК может регулироваться посредством модуляции длины поли(А)-хвоста. Ген Cnot1, кодирующий каркасный белок деаденилазного комплекса CCR4-NOT, экспрессируется на высоком уровне в супрахиазматическом ядре. Дефицит белка CNOT1 у мышей приводит к удлинению циркадного периода и изменению паттернов экспрессии мРНК и белков различных часовых генов, в основном Per2. Кроме того, CNOT1 связывается с мРНК Per2 через РНК-связывающий белок BRF1 (ZFP36L1), а нокдаун Brf1 приводит к повышению уровня экспрессии PER2 [58].
Большую роль в процессах посттранскрипционной регуляции играет альтернативный сплайсинг. Например, пре-мРНК гена Per2 альтернативно сплайсируется в соответствии с суточными ритмами, а вариант белка PER2S связан с нарушением организации ядрышка, которое действует как стимул, запускающий циркадные колебания [59]. Процессы посттранскрипционных модификаций хорошо изучены на млекопитающих, но не на зебраданио, что указывает на важность изучения особенностей циркадной регуляции рыб.
Посттрансляционные модификации и их роль в регуляции циркадных часов изучены более активно как у млекопитающих, так и у рыб (табл. 1). Одна из главных ролей в регуляции циркадных белков принадлежит казеинкиназам 1-го типа (СК-1) – семейству эволюционно консервативных киназ, регулирующих большое разнообразие внутриклеточных процессов эукариот [60]. Изоформы δ и ε принимают активное участие в фосфорилировании циркадных белков. CK-1δ играет ключевую роль в регуляции длины циркадного периода, тогда как CK-1ε играют второстепенную роль, что впоследствии было подтверждено на зебраданио с помощью селективных ингибиторов этих белков [61, 62].
Таблица 1.
Посттрансляционная регуляция циркадных белков c помощью CK-1, GSK-3 и SIRT1
| Регулятор | PER | CRY | REV-ERB | BMAL1 | CLOCK |
|---|---|---|---|---|---|
| CK-1 | (Ser487) Деградация [63] (Ser662) Стабилизация [64] |
Комплекс с PER, GAPVD1 Транслокация в ядро [69] |
– | – | Cовместно с CK-2 и PER2 Дестабилизация комплекса CLOCK:BMAL1 [42] |
| CК-1ε Ингибирование транспорта в ядро [67] | |||||
| GSK-3 | (Ser657)
Транслокация в ядро [72] |
(Ser557) Деградация [74] | (Ser55/Ser59) Стабилизация [73] |
(Ser17/Tyr21) Деградация [75] | (Ser-кластер) Деградация [76] |
| SIRT1 | Циркадная регуляция транскрипции генов | ||||
| Деградация [77] | – | – | Деацетилирование Снижение связывание с Е-боксом [79] Подавление активности в переферических тканях, но усиление в SCN [82] |
Образование комплекса усиление ацетилазной активности CLOCK [79] Подавление активности в переферических тканях, но усиление в SCN [82] |
|
CK-1 прежде всего являются регуляторами активности PER, поскольку фосфорилирование сайта Ser478 этого белка с помощью CK-1 необходимо для последующего полиубиквитинирования и деградации PER2 [63]. С другой стороны, фосфорилирование по сайту Ser662 необходимо для последующего множественного фосфорилирования CK-1, что стабилизирует PER2 [64]. Переключение между деградацией и стабилизацией зависит от температуры окружения, так как высокие температуры обычно увеличивают скорость биохимических реакций, в том числе и деградации PER2 [65]. При этом циркадные часы сохраняют примерно 24-часовой период независимо от температуры окружающей среды (явление, известное как температурная компенсация). Таким образом, циркадные часы компенсируют вызванное температурой увеличение скорости биохимических реакций. Например, при более высоких температурах среды происходит стабилизация PER2, тем самым компенсируя вызванное температурой ускорение его деградации [66].
Другая важная роль CK-1 – регулирование транспорта PER в ядро клеток. Например, CK-1ε ингибирует этот транспорт за счет фосфорилирования аминокислотных остатков, прилежащих к домену ядерной локализации белков PER [67]. В других работах CK-1, наоборот, способствуют транспорту через образование комплекса PER2 c CRY и GAPVD1 – фактора цитоплазматического переноса [68, 69]. Активность по отношению к PER2 определяется статусом специфического сайта Tyr347 на CK-1δ, который, вероятно, регулируется циклин-зависимыми киназами, а его мутация содействует деградации PER2 [70]. CK-1δ совместно с CK-2 и PER2 способствует фосфорилированию некоторых сайтов CLOCK в комплексе CLOCK:BMAL1, находящемся на промоторе активируемых генов. Гиперфосфорилированное состояние CLOCK определяет начало фазы репрессии транскрипции циркадных генов, когда связывание комплекса CLOCK:BMAL1 с промоторами ослабевает. Важно отметить, что проникновение CK-1δ напрямую требует PER и косвенно – CRY, поскольку они необходимы для проникновения PER в ядро [42].
Еще один важный регулятор циркадных ритмов – гликогенсинтазкиназы-3 (GSK-3), GSK-3α и β изоформы которых обнаружены в SCN, а их активность снижается в начале ночи и усиливается утром [71]. Фосфорилирование белков PER по сайту Ser657 с помощью GSK-3 способствует их транслокации в ядро [72]. Также GSK-3 способна стабилизировать REV-ERBα и способствует его транслокации в ядро, тем самым подавляя транскрипцию гена Bmal1 [73]. При этом GSK-3β играет большую роль в деградации белков. Так, фосфорилирование CRY2 по сайту Ser557, если имеется сайт p-Ser553, приводит к его протеосомной деградации [74], а фосфорилирование BMAL1 по сайтам Ser17/Tyr21 ведет к разрушению белка посредством убиквитинирования [75]. Помимо этого, GSK-3 принимает участие в регуляции особого серинового кластера белка CLOCK, названного фосфо-дегроном, что в условиях высокой активности GSK-3 ведет к деградации CLOCK [76].
NAD+-зависимая деацетилаза гистонов SIRT1 также играет важную роль в циркадной регуляции. Она необходима для обеспечения циркадной транскрипции Bmal1, Rorγ, Per2 и Cry1. Ритмичное связывание SIRT1 с комплексом CLOCK:BMAL1 способствует деацетилированию и деградации белка PER2 [77]. SIRT1 и PER2 составляют негативную реципрокную петлю, необходимую для модуляции циркадного ритма. Дефицит SIRT1 приводит к повышению экспрессии Per2, что еще больше подавляет экспрессию Sirt1, через взаимодействие PER2 с сайтом связывания CLOCK:BMAL1 комплекса на промоторе Sirt1 [78]. SIRT1 способен физически связываться с CLOCK, способствуя его ацетилазной активности, в том числе по отношению к сайту Lys537 в белке BMAL1. В то же время SIRT1 сам по себе способен деацетилировать BMAL1 в ответ на изменение концентрации NAD+, снижая его способность связываться с E-боксом, что указывает на роль клеточного метаболизма в циркадной регуляции. Также показано, что активность SIRT1, а не его мРНК, находится под контролем циркадных механизмов [79]. Ритмическая активность SIRT1 обусловлена колебательными паттернами уровней NAD+, а те, в свою очередь, вызваны активностью фермента никотинамидфосфорибозилтрансферазы (NAMPT), который положительно регулируется с помощью CLOCK:BMAL1. Активация SIRT1 через этот NAMPT-опосредованный путь подавляет активность CLOCK:BMAL1, тем самым формируя петлю отрицательной обратной связи [80, 81].
Несмотря на то, что в периферических тканях SIRT1 является негативным регулятором комплекса CLOCK:BMAL1, в SCN он способствует активации BMAL1 и CLOCK через рецептор PGC-1α, который является коактиватором транскрипции, связанным с метаболическими функциями клетки. Таким образом, регуляция суточных ритмов на уровне пострансляционных модификаций может происходить по-разному в разных тканях [82]. С помощью SIRT1 из вентромедиального гипоталамуса центральным часам передается информация о времени приема пищи, тем самым синхронизируя их с сигналами питания, что в очередной раз указывает на связь между метаболизмом и циркадным ритмом [83].
Среди менее изученных механизмов регуляции циркадных белков можно отметить роль протеинфосфатазы 4, которая напрямую взаимодействует с гетеродимером CLOCK:BMAL1 и снижает его активационный потенциал, тем самым, вероятно, участвуя в задержке активации генов-мишеней [84]. JUMONJIC и домен ARID, содержащий гистон-лизин-деметилазу 1a (JARID1a), образует комплекс с CLOCK:BMAL1, который рекрутируется на промотор Per2. Также JARID1a увеличивает ацетилирование гистонов путем ингибирования функции деацетилазы гистонов 1 и усиливает транскрипцию с помощью CLOCK:BMAL1. Истощение JARID1a в клетках млекопитающих снижает ацетилирование гистонов промотора Per2, ослабляет экспрессию циркадных генов и укорачивает период циркадных ритмов [85].
Белки PER2 также подвержены сумоилированию – процессу, который может изменять белок-белковые взаимодействия, субклеточную локализацию или активность белка и участвовать в регуляции транскрипции [86]. В сумоилировании PER2 принимают участие белки SUMO1 и SUMO2, а сайт Lys736 на PER2 необходим для взаимодействия с ними. SUMO2 облегчает взаимодействие PER2 с β-TrCP, что приводит к протеасомной деградации PER2. SUMO1, напротив, усиливает опосредованное CK-1 фосфорилирование сайта Ser662, ингибирует деградацию PER2 и повышает его функцию как супрессора транскрипции [87]. Таким образом, посттранскрипционные и посттрансляционные регуляторные механизмы в тканях в своем многообразии позволяют обеспечивать тонкую настройку циркадных ритмов организма.
СРЕДОВОЕ НАРУШЕНИЕ ЦИРКАДНЫХ РИТМОВ И ПОЛИМОРФИЗМЫ ЦИРКАДНЫХ ГЕНОВ КАК ФАКТОРЫ РАЗВИТИЯ ПСИХОПАТОЛОГИЙ
Как отмечалось ранее, свет крайне важен для настройки и синхронизации внутренних ритмов организма (рис. 1). ipRGC сетчатки в основном передают свет напрямую в SCN, однако некоторые популяции могут давать проекции в зоны мозга, регулирующие поведение. На мышах показано, что сигналы от ipRGC могут поступать в дорзальное перихабенулярное ядро, а оттуда в прилежащее ядро (NAc). Этот путь более активен в ночное время, что может опосредовать появление депрессивно-подобного фенотипа у мышей в условиях ночного освещения [19]. Нейроны из перихабенулярного региона дают проекции в другие области мозга, отвечающие за настроение (префронтальная кора, дорзомедиальный стриатум, NAc, латеральная хабенула), что объясняет поведенческие изменения в условиях нарушения светового режима [88]. Субпопуляция ipRGC, характеризующаяся низкой экспрессией гена Brn3b, может опосредовать вызванный ночным световым загрязнением когнитивный дефицит, не влияя на функции SCN как водителя ритма, что, по-видимому, отражает важность SCN для работы гиппокампа в процессе обучения, это подтверждалось более ранними исследованиями [88]. Все это доказывает, что свет, помимо настройки циркадных ритмов, может влиять на поведение и в определенных условиях нарушать его.
До того, как было введено повсеместное электрическое освещение, люди в ночное время подвергались минимальному освещению. Полная луна ясной ночью освещает окружение на 0.1–0.3 люкса (до 1.0 люкса в тропиках). Сегодня более 80% населения мира и 99% жителей США и Европы испытывают значительное ночное световое загрязнение (например, освещение типичного торгового центра составляет 10–20 люкс) [89]. И хотя экономические выгоды от внедрения электричества во все сферы производства привели к появлению ночных рабочих смен, работники ночной смены имеют повышенный риск развития различных заболеваний, в том числе и психических нарушений, таких как депрессия и тревожность [3]. С другой стороны, световую терапию используют для лечения несезонной депрессии и нормализации сна у пациентов с синдромом запаздывания времени сна (delayed sleep phase syndrome) [90–92].
Другой фактор – частые смены часовых поясов – может вызывать джетлаг-синдром, связанный с перестройкой организма под новые световые, социальные и пищевые ритмы. Поскольку этот синдром часто сопровождается бессонницей или гиперсомнией, он тоже может провоцировать усугубление психических расстройств [93, 94]. Предполагается, что циркадные ритмы человека обычно достаточно гибки, чтобы подстраиваться под смену часовых поясов и различные режимы освещения. Однако слабая чувствительность к свету приводит к снижению этой гибкости, а светотерапия может быть потенциальным подходом к решению проблем, связанных со сменной работой и частыми путешествиями [92]. Также предполагается, что слабая связь между системой AVP супрахиазматического ядра и HPA ослабляет гибкость суточных ритмов [95]. В свою очередь несоответствие между принудительной продолжительностью сна в рабочие и в выходные дни определяют как социальный джетлаг, который также влияет на психические функции, вызывая агрессию, расстройства настроения, ухудшение когнитивных показателей и употребление психоактивных веществ [96]. С другой стороны, поскольку не обнаружено прямой связи между социальным джетлагом и депрессивными симптомами, его влияние на поведение и здоровье человека требует более детального изучения [97]. Тем не менее роль внешних факторов среды в изменениях циркадной регуляции и психическом здоровье людей представляется существенной.
Молекулярные компоненты циркадных ритмов также могут быть вовлечены в различные патологии, включая психические расстройства человека. Пациенты с биполярным аффективным расстройством (БАР) и бессонницей часто несут полиморфизм в гене Clock (rs1801260C), вариант которого также связан с повышенной вероятностью попыток суицида у людей, подвергшихся стрессу, и с модификациями белого вещества мозга [98]. Полиморфизм гена Bmal1 вовлечен в регуляцию поведения и ассоциирован с БАР. Люди, несущие T/T генотип (rs7107287), имеют симптомы депрессии с циклотимическим темпераментом [99, 100], а полиморфизм rs2278749 этого гена коррелирует с увеличением массы тела, количеством принимаемой пищи и сезонным аффективным расстройством (САР) [101]. Два полиморфизма гена Per3 также вовлечены в развитие психопатологий: вариант rs228697 ассоциирован с повышенной тревожностью и, возможно, предрасположенностью к депрессии, а rs57875989 часто встречается у пациентов с депрессией [102, 103]. В Китае в популяции пациентов с клинической депрессий часто встречается С аллель гена Cry1 (rs2287161), полиморфизм которого связан с депрессивными эпизодами при биполярном расстройстве [104, 105] и депрессивными симптомами при болезни Паркинсона [106]. Полиморфизм в гене Cry2 (rs10838524) также ассоциирован с САР и дистимией [107–109].
Таким образом, выявляются два основных фактора, влияющих на развитие психопатологий: генетика циркадных компонентов и факторы внешней среды, обуславливающие циркадные процессы организма. Для создания экспериментальных моделей in vivo, исходя из перечисленных факторов, может активно использоваться генетическая модификация циркадных генов, моделирование влияния внешних факторов и совмещение этих двух подходов для изучения взаимодействия генетических и средовых факторов.
ЭКСПЕРИМЕНТАЛЬНЫЕ МОДЕЛИ ПСИХИЧЕСКИХ РАССТРОЙСТВ, СВЯЗАННЫЕ С НАРУШЕНИЕМ ЦИРКАДНОЙ РЕГУЛЯЦИИ
В настоящее время разработано множество экспериментальных моделей с нарушенным функционированием циркадных генов, показывающих признаки психических расстройств (табл. 2). На различных животных, преимущественно грызунах, изучаются потенциальные механизмы, вовлеченные в развитие патологических состояний, и методы их лечения. Например, мыши с мутацией в гене Clock (ClockΔ19) имеют фенотипические признаки, сходные с симптоматикой БАР, в т.ч. снижение тревожности и сокращение времени сна, гиперактивность и повышенный ответ на подкрепление [110]. У них наблюдается снижение опосредованного AMPA-рецепторами возбуждения нейронов в NАc из-за гиперполяризации постсинаптической мембраны в состоянии покоя, что связано со снижением количества белка GRIA1 на поверхности мембраны и нарушением ритма его синтеза. Помимо этого, ClockΔ19-мыши характеризуются повышенной активностью дофаминовых нейронов в вентральной области покрышки (VTA) и повышенным дофаминовым сигналингом в NAc [110]. Нарушения поведения в этой модели нормализуются литием, способным ингибировать активность GSK-3, и блокаторами CK-1δ/ε [111, 112]. Эта же мутация, но специфично вызванная в вентральной покрышечной области (ClockΔ19-VTA), приводит к появлению смешанного состояния маниакально-подобного и депрессивно-подобного поведения, изменению циркадного ритма двигательной активности и повышенной активности нейронов в данной области, а также изменению экспрессии генов ионных каналов и генов, связанных с метаболизмом дофамина [113].
Таблица 2.
Экспериментальные модели нарушения циркадных ритмов
| Животное | Модель | Поведенческий фенотип | Затронутые механизмы | Источник | |
|---|---|---|---|---|---|
| Модели нарушения циркадных генов | |||||
| Мышь (Mus musculus) |
ClockΔ19 | Маниакально-подобное поведение:
гиперактивность; снижение тревожности и времени сна; повышенный ответ на вознаграждение |
Повышение активности дофаминовых нейронов в VTA и дофаминового сигналинга в NAc; гиперполяризованность мембраны шипиков NAc в состоянии покоя; снижение количества белка GRIA1 на поверхности мембраны; нарушения поведения нормализуются литием и блокаторами КK-1δ/ε |
[110–112] | |
| ClockΔ19-VTA | Маниакально- и депрессивно-подобное
поведение: гиперактивность; снижение тревожности; неподвижность в тесте принудительного плавания; Изменение циркадного ритма локомоторной активности |
Повышенная активность дофаминергических нейронов VTA; изменение экспрессии генов ионных каналов и метаболизма дофамина |
[113] | ||
| SCN-Bmal1-KD | Депрессивно-подобный фенотип | Снижение массы тела; аномальный циркадный ритм кортикостерона |
[115] | ||
| Bmal1-KO | Депрессивно-подобный фенотип: ангедония (снижение предпочтения сахарозы) |
Гипокортицизм; нарушение реакции надпочечников на АКТГ; Снижение транскрипции генов транспорта холестерина в клетках коры надпочечников |
[116] | ||
| Rev-erbα-KO | Дефицит краткосрочной, долгосрочной и контекстной памяти; нарушение гнездового поведения |
Усиление оборота дофамина в гиппокампе | [117] | ||
| NAc-Per1/2-KD | Тревожный фенотип | – | [118] | ||
| Per3-KO (hPer3-P415A/H417R) |
Нарушение ритмов локомоторной активности; депрессивно-подобный фенотип |
Нарушение стабильности PER1, PER2 | [120] | ||
| Cry1-KO | Тревожный фенотип | – | [122] | ||
| Cry2-KO | |||||
| Cry1/2-KO | Снижение спонтанной локомоции; нарушение памяти распознавания, при сохранении памяти о страхе, тревожность; высокая чувствительность к препаратам-психостимуляторам |
Снижение уровня фосфорилирования белка ERK в MSN | |||
| Макака крабоед (Macaca fascicularis) |
Bmal1-KO | Снижение количества сна и повышение активности в ночное время; тревожно-, депрессивно- и шизофрено-подобный фенотип |
Аритмичность колебания уровней гормонов; снижение экспрессии провоспалительных цитокинов и уровня кортизола в крови |
[114] | |
| Крыса (Rattus norvegicus domestica) |
Per2-KD-LHb | Депрессивно-подобное поведение (преимущественно в ночное время) | – | [119] | |
| Зебраданио (Danio rerio) |
per1b-KO | Гиперактивность и импульсивность; дефицит обучения и памяти (поведение, подобное дефициту внимания) |
Снижение уровня дофамина в теле мальков и мозге взрослых рыб; повышение экспрессии генов метаболизма дофамина; восьмичасовая задержка экспрессии rev-erbα и последующая задержка экспрессии тирозингидроксилазы |
[121] | |
| Модели внешнего воздействия | |||||
| Мышь (Mus musculus) |
Тусклый свет в ночное время (3 ночи) | Депрессивно-подобный фенотип | Нейровоспаление; снижение плотности дендритных шипиков в гиппокампе; изменения экспрессии циркадных генов в гиппокампе; |
[124] | |
| 20-часовой цикл | Изменение циркадной регуляции температуры тела; снижение когнитивной гибкости; изменения эмоциональности |
Снижение массы тела и длины дендритных отростков в префронтальной коре; повышение концентрации инсулина и лептина в крови |
[127] | ||
| Лишение сна | Симптомы мании | Нарушение функционирования HPA; повышение концентрации цитокинов в мозге и сыворотке крови; возвращение к нормальному фенотипу с помощью лития |
[128] | ||
| СХСП | Депрессивно-подобный и тревожный фенотип; изменение архитектуры сна, циркадного ритма двигательной активности и температуры тела |
Изменение уровней мРНК Per2 в зонах мозга, связанных с эмоциями и мотивацией; антагонисты κ-опиоидных рецепторов перед воздействием СХСП смягчали последующие патологические изменения |
[129, 130] | ||
| Тревожный фенотип | Снижение экспрессии генов Per1 и Per2 в NAc; восстановление антидепрессантами | [118, 130] | |||
| Крыса (Rattus norvegicus domestica) |
Непрерывное дневное освещение (8 нед.) | Тревожно- и депрессивно-подобное поведение; изменение ритмов двигательной активности |
Нарушение ритмичности выработки мелатонина и кортикостерона; снижение нейрональной активности клеток SCN |
[123] | |
| Пре- и постнатальное постоянное освещение | Тревожный фенотип | Не обнаружено нарушений во взрослом возрасте | [125] | ||
| Стресс раннего периода жизни | – | Повышенная концентрация глюкокортикоидов в плазме крови; сдвиг ритма экспрессии Bmal1 в СХЯ |
[132] | ||
| Сибирский хомячок (Phodopus sungorus) | Тусклый свет в ночное время (1 нед.) | Незначительные изменения двигательной активности | Нарушение суточных колебаний концентрации кортизола; изменение паттернов экспрессии белков часовых генов в SCN и гиппокампе |
[126] | |
| Модели взаимодействия генетических и средовых факторов | |||||
| Мышь (Mus musculus) |
Bmal1-KO + повторяющийся стресс обездвиживания |
Слабо восприимчивы к стрессу | Отсутствие изменений в уровне кортикостерона при повышении концентрации АКТГ в крови | [116] | |
| Per3-KO (hPer3-P415A/H417R) + 4:20 цикл света/темноты |
Депрессивно-подобное поведение | – | [120] | ||
| Макака-крабоед (Macaca fascicularis) |
Bmal1-KO + постоянное освещение |
Усугубление поведенческого фенотипа у нокаутов | [114] | ||
Нокаут Bmal1 (Bmal1-KO) у макак-крабоедов (Macaca fascicularis) снижает количество сна и повышает активность в ночное время, понижает и нарушает ритмичность выработки гормонов, таких как мелатонин, тестостерон и дегидроэпиандростерон [114]. Параллельно наблюдается усиленная экспрессия провоспалительных цитокинов в крови и наличие системного воспаления, а также признаки тревожности и депрессивно-подобного поведения, связанного с повышенным уровнем кортизола в крови, а также нарушения сенсорной обработки, что характерно для шизофрено-подобного поведения. Эти поведенческие признаки усугублялись в условиях постоянного освещения [114]. SCN-специфический нокдаун Bmal1 (SCN-Bmal1-KD) приводит к появлению депрессивно-подобного фенотипа у мышей, увеличению веса тела, аномальному циркадному ритму кортикостерона и его слабому повышению в ответ на стресс [115]. Мыши, нокаутные по Bmal1 (Bmal1-KO), характеризуются ангедонией (сниженным предпочтением сахарозы), что является признаком депрессивно-подобного поведения, низким уровнем кортикостерона и нарушением ответа надпочечников на адренокортикотропный гормон (АКТГ) в связи с подавлением транскрипции генов, участвующих в транспорте холестерина в клетках надпочечников [116]. Все это приводит к отсутствию выраженного поведенческого ответа на острый и субхронический стресс на фоне стабильного уровня кортикостерона в крови до и после воздействия стресса, тогда как повышение концентрации АКТГ в ответ на стресс явно выражено [116].
Мыши с нокаутом Rev-erbα (Rev-erbα-KO) обладают дефицитом кратковременной, долговременной и контекстной памяти, а также демонстрируют нарушение способности строить гнезда, что отражает нарушение функций гиппокампа и усиление оборота дофамина в нем [117]. Одновременный нокдаун генов Per1/2 в прилежащем ядре мышей (NAc-Per1/2-KD) приводит к появлению у них тревожного фенотипа в большей мере, чем нокдаун Per1 или Per2 по отдельности [118]. Нокдаун Per2 в латеральной хабенуле крыс (Per2-KD-LHb) приводит к появлению депрессивно-подобного поведения, преимущественно в ночное время [119]. Трансгенные мыши с мутацией гена Per3 (hPer3-P415A/H417R), обуславливающей развитие у людей синдрома продвинутой фазы сна (advanced sleep phase syndrome), показывают нарушение ритмов локомоторной активности и депрессивно-подобный фенотип, что связано с нарушением стабильности белка PER3 и ведет к ослаблению стабильности белков PER1 и PER2 [120]. Кроме того, эти животные в условиях короткого фотопериода (4:20 цикл света/темноты) показывают признаки депрессивно-подобного поведения [120]. При этом нокаут гена per1b у зебраданио вызывает гиперактивность, импульсивность, дефицит обучения и памяти и поведение, подобное дефициту внимания, снижение уровня дофамина в теле мальков и мозге взрослых рыб, что соотносится с данными, полученными на мышах и с симптоматикой людей с синдромом дефицита внимания и гиперреактивности [121]. У этих рыб обнаружена восьмичасовая задержка экспрессии rev-erbα, что влияет на задержку экспрессии гена тирозингидроксилазы [121].
Мыши с нокаутом генов Cry1/2 (Cry1/2-KO) характеризуются снижением спонтанной двигательной активности и нарушением распознавания при сохранении памяти о страхе. Двойные мутанты проявляют повышенную тревожность, а нокаут по генам Cry1 и Cry2 в отдельности приводит к менее выраженному тревожному фенотипу [122]. Мыши Cry1/2 также демонстрируют большую чувствительность к препаратам-психостимуляторам, что связано со сниженным уровнем фосфорилирования белка ERK в средних шипиковых нейронах (medium spiny neurons, MSN) [122].
Влияние внешних факторов на циркадные ритмы и поведение также широко исследуется с использованием моделей in vivo. Крысы, подвергшиеся непрерывному дневному освещению в течение 8 недель, проявляют признаки тревожного и депрессивно-подобного поведения, у этих животных наблюдается изменение ритмов двигательной активности, выработки мелатонина и кортикостерона, снижение нейрональной активности в SCN [123]. Тусклый свет в течение трех ночей приводит к появлению депрессивно-подобного фенотипа у мышей, нейровоспалению, снижению плотности дендритных шипиков и изменению экспрессии циркадных генов в гиппокампе [124]. Пре- и постнатальное трехнедельное воздействие тусклым светом в ночное время на крыс может приводить к появлению тревожности во взрослом возрасте [125]. У сибирских хомячков (Phodopus sungorus) постоянное освещение в течение недели приводило к нарушению суточных колебаний концентрации кортизола и изменению паттернов экспрессии белков часовых генов в SCN и гиппокампе [126].
Другой подход к моделированию циркадных и поведенческих нарушений – изменение длины цикла света/темноты. Содержание мышей в 20-часовом цикле приводит к снижению длины дендритных отростков в префронтальной коре, изменению циркадной регуляции температуры тела, увеличению массы тела, повышению концентрации гормонов инсулина и лептина в крови, снижению когнитивной гибкости и изменениям эмоциональности [127]. Лишение сна при сохранении нормального цикла света/темноты приводит к появлению симптомов мании у мышей, нарушению функционирования HPA, вызывает окислительный стресс, повышение концентрации цитокинов в мозге и сыворотке крови (однако эти последствия могут быть нормализованы литием) [128].
Стресс хронического социального поражения (СХСП) широко используется как модель для создания тревожного и депрессивно-подобного поведения у грызунов [129]. СХСП оказывает влияние на архитектуру сна и динамику уровней мРНК Per2 в зонах мозга, связанных с эмоциями и мотивацией [129, 130] (экспрессия Per1 и Per2 снижена в NAc у мышей после десятидневного СХСП, но возвращается к нормальному состоянию после лечения антидепрессантами) [118]. При этом СХСП не изменяет циркадные ритмы в SCN, что, вероятно, связано с малым количеством рецепторов к глюкокортикоидам в клетках данного ядра [131]. У новорожденных крысят присутствует достаточное количество этих рецепторов в SCN, из-за чего стресс раннего периода жизни (до 6- дневного возраста) оказывает влияние на фазу генной экспрессии SCN [132].
В настоящее время имеется немного исследований, посвященных взаимодействию генетических и средовых факторов циркадной регуляции, его роли в развитии психопатологий. Количество работ, исследующих патологическое поведение зебраданио в контексте нарушения циркадной регуляции и психопатологии, также незначительно. В то же время зебраданио обладают рядом важных характеристик, позволяющих использовать их как перспективную модель психопатологий, а также для изучения механизмов регуляции циркадных ритмов. Одним из таких преимуществ является то, что состояния бодрствования и покоя у рыб легко определяются поведенческими критериями. Поэтому фенотипы повышенной или пониженной суточной активности и изменение длительности периодов бодрствования и покоя можно использовать в качестве маркеров поведенческих нарушений. Зебраданио также удобны и для фармакологического скрининга препаратов, используемых для улучшения качества сна и лечения психопатологий [133], например, мелатонина [133, 134]. Кроме того, консервативность циркадных процессов и указанные ранее сходства в регуляции молекулярных компонентов циркадных ритмов делают зебраданио удобным объектом для трансляционной циркадной биологии в целом.
ЗАКЛЮЧЕНИЕ
Циркадные ритмы – важный и неотъемлемый фактор жизни, от которого во многом зависит психическое здоровье человека. Их нарушения как со стороны генетических, так и факторов внешней среды, могут приводить к развитию серьезных психопатологий и расстройств поведения. Регуляция биоритмов в организме осуществляется на различных уровнях, в том числе на уровне посттранскрипционной и посттрансляционной обработки продуктов циркадных генов, поэтому понимание этих механизмов в экспериментальных моделях может помочь в разработке терапевтических агентов, нацеленных на разные стадии этого процесса. Это также необходимо и для выработки профилактических мер, способствующих предотвращению поведенческих нарушений, вызванных аномалиями циркадной регуляции.
Создание новых экспериментальных моделей психопатологий с упором на циркадные функции также является важным направлением научно-исследовательской деятельности, в том числе с использованием новых перспективных модельных систем (например, зебраданио). Так, например, потенциал для скрининга физиологически активных веществ на зебраданио в десятки или сотни раз превышает возможности подобных скринов на грызунах, что позволяет не только выявлять принципиально новые регуляторы циркадных ритмов в ходе высокопроизводительного тестирования на рыбах, но и проводить перепрофилирование лекарственных препаратов (drug repurposing) на предмет возможной модуляции циркадных ритмов препаратами, уже одобренными для клинического применения в других целях. В целом, несмотря на большой объем данных, посвященных центральной регуляции циркадных ритмов, остается множество открытых вопросов (табл. 3), которые могут служить фундаментом для будущих научных работ по данной теме.
Таблица 3.
Перечень малоизученных вопросов по проблемам центральной регуляции циркадных ритмов
| 1. | Функции глубинных фоторецепторов рыб в циркадной регуляции и регуляции поведения; |
| 2. | Роль генетических различий (гомология геномов ~70%) между человеком и зебраданио в регуляции циркадных ритмов; |
| 3. | Роль эпигенетических механизмов ЦНС в регуляции циркадных ритмов человека и зебраданио; |
| 4. | Установка ритмов экспрессии циркадных генов в периферических тканях; |
| 5. | Пострансляционная и постранскрипционная обработка продуктов циркадных генов у рыб; |
| 6. | Влияния социального джетлага на поведение; |
| 7. | Взаимодействия мутаций в циркадных генах и внешних факторов, и роль этого взаимодействия в развитии психопатологий; |
| 8. | Поведение зебраданио в контексте нарушения циркадной регуляции и психопатологии. |
Список литературы
Bhadra U, Thakkar N, Das P, Pal Bhadra M (2017) Evolution of circadian rhythms: from bacteria to human. Sleep Med 35: 49–61. https://doi.org/10.1016/j.sleep.2017.04.008
Herzog ED, Hermanstyne T, Smyllie NJ, Hastings MH (2017) Regulating the Suprachiasmatic Nucleus (SCN) Circadian Clockwork: Interplay between Cell-Autonomous and Circuit-Level Mechanisms. Cold Spring Harb Perspect Biol 9: a027706. https://doi.org/10.1101/cshperspect.a027706
Lee A, Myung S-K, Cho JJ, Jung Y-J, Yoon JL, Kim MY (2017) Night Shift Work and Risk of Depression: Meta-analysis of Observational Studies. J Korean Med Sci 32: 1091. https://doi.org/10.3346/jkms.2017.32.7.1091
Lu Z, Klein-Cardeña K, Lee S, Antonsen TM, Girvan M, Ott E (2016) Resynchronization of circadian oscillators and the east-west asymmetry of jet-lag. Chaos An Interdiscip J Nonlinear Sci 26: 094811. https://doi.org/10.1063/1.4954275
Haesemeyer M, Schier AF (2015) The study of psychiatric disease genes and drugs in zebrafish. Curr Opin Neurobiol 30: 122–130. https://doi.org/10.1016/j.conb.2014.12.002
Fontana BD, Mezzomo NJ, Kalueff AV, Rosemberg DB (2018) The developing utility of zebrafish models of neurological and neuropsychiatric disorders: A critical review. Exp Neurol 299: 157–171. https://doi.org/10.1016/j.expneurol.2017.10.004
Aranda ML, Schmidt TM (2021) Diversity of intrinsically photosensitive retinal ganglion cells: circuits and functions. Cell Mol Life Sci 78: 889–907. https://doi.org/10.1007/s00018-020-03641-5
Do MTH (2019) Melanopsin and the Intrinsically Photosensitive Retinal Ganglion Cells: Biophysics to Behavior. Neuron 104: 205–226. https://doi.org/10.1016/j.neuron.2019.07.016
Davies WIL, Zheng L, Hughes S, Tamai TK, Turton M, Halford S, Foster RG, Whitmore D, Hankins MW (2011) Functional diversity of melanopsins and their global expression in the teleost retina. Cell Mol Life Sci 68: 4115–4132. https://doi.org/10.1007/s00018-011-0785-4
Matsuyama T, Yamashita T, Imamoto Y, Shichida Y (2012) Photochemical Properties of Mammalian Melanopsin. Biochemistry 51: 5454–5462. https://doi.org/10.1021/bi3004999
Walmsley L, Hanna L, Mouland J, Martial F, West A, Smedley AR, Bechtold DA, Webb AR, Lucas RJ, Brown TM (2015) Colour As a Signal for Entraining the Mammalian Circadian Clock. PLoS Biol 13: e1002127. https://doi.org/10.1371/journal.pbio.1002127
Steindal IAF, Whitmore D (2020) Zebrafish Circadian Clock Entrainment and the Importance of Broad Spectral Light Sensitivity. Front Physiol 11. https://doi.org/10.3389/fphys.2020.01002
Jones JR, Simon T, Lones L, Herzog ED (2018) SCN VIP Neurons Are Essential for Normal Light-Mediated Resetting of the Circadian System. J Neurosci 38: 7986–7995. https://doi.org/10.1523/JNEUROSCI.1322-18.2018
Ono D, Honma K, Yanagawa Y, Yamanaka A, Honma S (2018) Role of GABA in the regulation of the central circadian clock of the suprachiasmatic nucleus. J Physiol Sci 68: 333–343. https://doi.org/10.1007/s12576-018-0604-x
Ukraintseva YV, Kovalzon VM (2016) Circadian regulation and its disorders in Parkinson’s disease patients. Part 2: Experimental models, alpha-synuclein, and melatonin. Hum Physiol 42: 559–570. https://doi.org/10.1134/S0362119716050170
Mazuski C, Abel JH, Chen SP, Hermanstyne TO, Jones JR, Simon T, Doyle FJ, Herzog ED (2018) Entrainment of Circadian Rhythms Depends on Firing Rates and Neuropeptide Release of VIP SCN Neurons. Neuron 99: 555–563.e5. https://doi.org/10.1016/j.neuron.2018.06.029
Kalsbeek A, Fliers E, Hofman MA, Swaab DF, Buijs RM (2010) Vasopressin and the Output of the Hypothalamic Biological Clock. J Neuroendocrinol 22: 362–372. https://doi.org/10.1111/j.1365-2826.2010.01956.x
Mieda M, Ono D, Hasegawa E, Okamoto H, Honma K, Honma S, Sakurai T (2015) Cellular Clocks in AVP Neurons of the SCN Are Critical for Interneuronal Coupling Regulating Circadian Behavior Rhythm. Neuron 85: 1103–1116. https://doi.org/10.1016/j.neuron.2015.02.005
An K, Zhao H, Miao Y, Xu Q, Li Y-F, Ma Y-Q, Shi Y-M, Shen J-W, Meng J-J, Yao Y-G, Zhang Z, Chen J-T, Bao J, Zhang M, Xue T (2020) A circadian rhythm-gated subcortical pathway for nighttime-light-induced depressive-like behaviors in mice. Nat Neurosci 23: 869–880. https://doi.org/10.1038/s41593-020-0640-8
Campos LMG, Cruz-Rizzolo RJ, Watanabe IS, Pinato L, Nogueira MI (2014) Efferent projections of the suprachiasmatic nucleus based on the distribution of vasoactive intestinal peptide (VIP) and arginine vasopressin (AVP) immunoreactive fibers in the hypothalamus of Sapajus apella. J Chem Neuroanat 57–58: 42–53. https://doi.org/10.1016/j.jchemneu.2014.03.004
Malek ZS, Labban LM (2021) Photoperiod regulates the daily profiles of tryptophan hydroxylase-2 gene expression the raphe nuclei of rats. Int J Neurosci 131: 1155–1161. https://doi.org/10.1080/00207454.2020.1782903
Buijs FN, Guzmán-Ruiz M, León-Mercado L, Basualdo MC, Escobar C, Kalsbeek A, Buijs RM (2017) Suprachiasmatic Nucleus Interaction with the Arcuate Nucleus; Essential for Organizing Physiological Rhythms. Eneuro 4: ENEURO.0028-17.2017. https://doi.org/10.1523/ENEURO.0028-17.2017
Ni R-J, Shu Y-M, Luo P-H, Zhou J-N (2021) Whole-brain mapping of afferent projections to the suprachiasmatic nucleus of the tree shrew. Tissue Cell 73: 101620. https://doi.org/10.1016/j.tice.2021.101620
Brancaccio M, Patton AP, Chesham JE, Maywood ES, Hastings MH (2017) Astrocytes Control Circadian Timekeeping in the Suprachiasmatic Nucleus via Glutamatergic Signaling. Neuron 93: 1420–1435.e5. https://doi.org/10.1016/j.neuron.2017.02.030
Ortinski P, Reissner K, Turner J, Anderson TA, Scimemi A (2022) Control of complex behavior by astrocytes and microglia. Neurosci Biobehav Rev 137: 104651. https://doi.org/10.1016/j.neubiorev.2022.104651
Hang CY, Kitahashi T, Parhar IS (2014) Localization and characterization of val-opsin isoform-expressing cells in the brain of adult zebrafish. J Comp Neurol 522: 3847–3860. https://doi.org/10.1002/cne.23645
Moore HA, Whitmore D (2014) Circadian Rhythmicity and Light Sensitivity of the Zebrafish Brain. PLoS One 9: e86176. https://doi.org/10.1371/journal.pone.0086176
Horstick EJ, Bayleyen Y, Sinclair JL, Burgess HA (2017) Search strategy is regulated by somatostatin signaling and deep brain photoreceptors in zebrafish. BMC Biol 15: 4. https://doi.org/10.1186/s12915-016-0346-2
Fernandes AM, Fero K, Arrenberg AB, Bergeron SA, Driever W, Burgess HA (2012) Deep Brain Photoreceptors Control Light-Seeking Behavior in Zebrafish Larvae. Curr Biol 22: 2042–2047. https://doi.org/10.1016/j.cub.2012.08.016
Hang CY, Kitahashi T, Parhar IS (2015) Brain area-specific diurnal and photic regulation of val-opsinA and val-opsinB genes in the zebrafish. J Neurochem 133: 501–510. https://doi.org/10.1111/jnc.13084
Ben-Moshe Livne Z, Alon S, Vallone D, Bayleyen Y, Tovin A, Shainer I, Nisembaum LG, Aviram I, Smadja-Storz S, Fuentes M, Falcón J, Eisenberg E, Klein DC, Burgess HA, Foulkes NS, Gothilf Y (2016) Genetically Blocking the Zebrafish Pineal Clock Affects Circadian Behavior. PLoS Genet 12: e1006445. https://doi.org/10.1371/journal.pgen.1006445
Krylov VV, Izvekov EI, Pavlova VV, Pankova NA, Osipova EA (2021) Circadian rhythms in zebrafish (Danio rerio) behavour and the sources of their variability. Biol Rev 96: 785–797. https://doi.org/10.1111/brv.12678
Landgraf D, Achten C, Dallmann F, Oster H (2015) Embryonic development and maternal regulation of murine circadian clock function. Chronobiol Int 32: 416–427. https://doi.org/10.3109/07420528.2014.986576
Laurà R, Magnoli D, Zichichi R, Guerrera MC, De Carlos F, Suárez AÁ, Abbate F, Ciriaco E, Vega JA, Germanà A (2012) The photoreceptive cells of the pineal gland in adult zebrafish (Danio rerio). Microsc Res Tech 75: 359–366. https://doi.org/10.1002/jemt.21064
Yáñez J, Busch J, Anadón R, Meissl H (2009) Pineal projections in the zebrafish (Danio rerio): overlap with retinal and cerebellar projections. Neuroscience 164: 1712–1720. https://doi.org/10.1016/j.neuroscience.2009.09.043
Sato F, Kohsaka A, Bhawal U, Muragaki Y (2018) Potential Roles of Dec and Bmal1 Genes in Interconnecting Circadian Clock and Energy Metabolism. Int J Mol Sci 19: 781. https://doi.org/10.3390/ijms19030781
Gompf HS, Fuller PM, Hattar S, Saper CB, Lu J (2015) Impaired Circadian Photosensitivity in Mice Lacking Glutamate Transmission from Retinal Melanopsin Cells. J Biol Rhythms 30: 35–41. https://doi.org/10.1177/0748730414561545
Koyanagi S, Hamdan AM, Horiguchi M, Kusunose N, Okamoto A, Matsunaga N, Ohdo S (2011) cAMP-response Element (CRE)-mediated Transcription by Activating Transcription Factor-4 (ATF4) Is Essential for Circadian Expression of the Period 2 Gene. J Biol Chem 286: 32416–32423. https://doi.org/10.1074/jbc.M111.258970
Lindberg PT, Mitchell JW, Burgoon PW, Beaulé C, Weihe E, Schäfer MK-H, Eiden LE, Jiang SZ, Gillette MU (2019) Pituitary Adenylate Cyclase-Activating Peptide (PACAP)-Glutamate Co-transmission Drives Circadian Phase-Advancing Responses to Intrinsically Photosensitive Retinal Ganglion Cell Projections by Suprachiasmatic Nucleus. Front Neurosci 13: 1281. https://doi.org/10.3389/fnins.2019.01281
Yu W, Nomura M, Ikeda M (2002) Interactivating Feedback Loops within the Mammalian Clock: BMAL1 Is Negatively Autoregulated and Upregulated by CRY1, CRY2, and PER2. Biochem Biophys Res Commun 290: 933–941. https://doi.org/10.1006/bbrc.2001.6300
Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U (2010) The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev 24: 345–357.https://doi.org/10.1101/gad.564110
Cao X, Yang Y, Selby CP, Liu Z, Sancar A (2021) Molecular mechanism of the repressive phase of the mammalian circadian clock. Proc Natl Acad Sci U S A 118. https://doi.org/10.1073/pnas.2021174118
Bozek K, Relógio A, Kielbasa SM, Heine M, Dame C, Kramer A, Herzel H (2009) Regulation of Clock-Controlled Genes in Mammals. PLoS One 4: e4882. https://doi.org/10.1371/journal.pone.0004882
Crumbley C, Burris TP (2011) Direct Regulation of CLOCK Expression by REV-ERB. PLoS One 6: e17290. https://doi.org/10.1371/journal.pone.0017290
Takeda Y, Jothi R, Birault V, Jetten AM (2012) RORγ directly regulates the circadian expression of clock genes and downstream targets in vivo. Nucleic Acids Res 40: 8519–8535. https://doi.org/10.1093/nar/gks630
Ye R, Selby CP, Chiou Y-Y, Ozkan-Dagliyan I, Gaddameedhi S, Sancar A (2014) Dual modes of CLOCK:BMAL1 inhibition mediated by Cryptochrome and Period proteins in the mammalian circadian clock. Genes Dev 28: 1989–1998. https://doi.org/10.1101/gad.249417.114
Kwon I, Lee J, Chang SH, Jung NC, Lee BJ, Son GH, Kim K, Lee KH (2006) BMAL1 Shuttling Controls Transactivation and Degradation of the CLOCK/BMAL1 Heterodimer. Mol Cell Biol 26: 7318–7330. https://doi.org/10.1128/MCB.00337-06
Ono D, Honma K, Schmal C, Takumi T, Kawamoto T, Fujimoto K, Kato Y, Honma S (2021) CHRONO and DEC1/DEC2 compensate for lack of CRY1/CRY2 in expression of coherent circadian rhythm but not in generation of circadian oscillation in the neonatal mouse SCN. Sci Rep 11: 19240. https://doi.org/10.1038/s41598-021-98532-5
Li Y, Li G, Wang H, Du J, Yan J (2013) Analysis of a Gene Regulatory Cascade Mediating Circadian Rhythm in Zebrafish. PLoS Comput Biol 9: e1002940. https://doi.org/10.1371/journal.pcbi.1002940
West AC, Iversen M, Jørgensen EH, Sandve SR, Hazlerigg DG, Wood SH (2020) Diversified regulation of circadian clock gene expression following whole genome duplication. PLoS Genet 16: e1009097. https://doi.org/10.1371/journal.pgen.1009097
Wang M, Zhong Z, Zhong Y, Zhang W, Wang H (2015) The Zebrafish Period2 Protein Positively Regulates the Circadian Clock through Mediation of Retinoic Acid Receptor (RAR)-related Orphan Receptor α (Rorα). J Biol Chem 290: 4367–4382. https://doi.org/10.1074/jbc.M114.605022
Sloin HE, Ruggiero G, Rubinstein A, Smadja Storz S, Foulkes NS, Gothilf Y (2018) Interactions between the circadian clock and TGF-β signaling pathway in zebrafish. PLoS One 13: e0199777. https://doi.org/10.1371/journal.pone.0199777
Finger A-M, Jäschke S, del Olmo M, Hurwitz R, Granada AE, Herzel H, Kramer A (2021) Intercellular coupling between peripheral circadian oscillators by TGF-β signaling. Sci Adv 7. https://doi.org/10.1126/sciadv.abg5174
Mure LS, Le HD, Benegiamo G, Chang MW, Rios L, Jillani N, Ngotho M, Kariuki T, Dkhissi-Benyahya O, Cooper HM, Panda S (2018) Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science (80): 359. https://doi.org/10.1126/science.aao0318
Koike N, Yoo S-H, Huang H-C, Kumar V, Lee C, Kim T-K, Takahashi JS (2012) Transcriptional Architecture and Chromatin Landscape of the Core Circadian Clock in Mammals. Science (80) 338: 349–354. https://doi.org/10.1126/science.1226339
Terajima H, Yoshitane H, Ozaki H, Suzuki Y, Shimba S, Kuroda S, Iwasaki W, Fukada Y (2017) ADARB1 catalyzes circadian A-to-I editing and regulates RNA rhythm. Nat Genet 49: 146–151. https://doi.org/10.1038/ng.3731
Fustin J-M, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, Okamura H (2013) RNA-Methylation-Dependent RNA Processing Controls the Speed of the Circadian Clock. Cell 155: 793–806. https://doi.org/10.1016/j.cell.2013.10.026
Mohamed HMA, Takahashi A, Nishijima S, Adachi S, Murai I, Okamura H, Yamamoto T (2022) CNOT1 regulates circadian behaviour through Per2 mRNA decay in a deadenylation-dependent manner. RNA Biol 19: 703–718. https://doi.org/10.1080/15476286.2022.2071026
Avitabile D, Genovese L, Ponti D, Ranieri D, Raffa S, Calogero A, Torrisi MR (2014) Nucleolar localization and circadian regulation of Per2S, a novel splicing variant of the Period 2 gene. Cell Mol Life Sci 71: 2547–2559. https://doi.org/10.1007/s00018-013-1503-1
Fan J-Y, Preuss F, Muskus MJ, Bjes ES, Price JL (2009) Drosophila and Vertebrate Casein Kinase Iδ Exhibits Evolutionary Conservation of Circadian Function. Genetics 181: 139–152. https://doi.org/10.1534/genetics.108.094805
Etchegaray J-P, Machida KK, Noton E, Constance CM, Dallmann R, Di Napoli MN, DeBruyne JP, Lambert CM, Yu EA, Reppert SM, Weaver DR (2009) Casein Kinase 1 Delta Regulates the Pace of the Mammalian Circadian Clock. Mol Cell Biol 29: 3853–3866. https://doi.org/10.1128/MCB.00338-09
Smadja Storz S, Tovin A, Mracek P, Alon S, Foulkes NS, Gothilf Y (2013) Casein Kinase 1δ Activity: A Key Element in the Zebrafish Circadian Timing System. PLoS One 8: e54189. https://doi.org/10.1371/journal.pone.0054189
Masuda S, Narasimamurthy R, Yoshitane H, Kim JK, Fukada Y, Virshup DM (2020) Mutation of a PER2 phosphodegron perturbs the circadian phosphoswitch. Proc Natl Acad Sci U S A 117: 10888–10896. https://doi.org/10.1073/pnas.2000266117
Shanware NP, Hutchinson JA, Kim SH, Zhan L, Bowler MJ, Tibbetts RS (2011) Casein Kinase 1-dependent Phosphorylation of Familial Advanced Sleep Phase Syndrome-associated Residues Controls PERIOD 2 Stability. J Biol Chem 286: 12766–12774. https://doi.org/10.1074/jbc.M111.224014
Zhou M, Kim JK, Eng GWL, Forger DB, Virshup DM (2015) A Period2 Phosphoswitch Regulates and Temperature Compensates Circadian Period. Mol Cell 60: 77–88. https://doi.org/10.1016/j.molcel.2015.08.022
Narasimamurthy R, Virshup DM (2017) Molecular Mechanisms Regulating Temperature Compensation of the Circadian Clock. Front Neurol 8. https://doi.org/10.3389/fneur.2017.00161
Vielhaber EL, Duricka D, Ullman KS, Virshup DM (2001) Nuclear Export of Mammalian PERI-OD Proteins. J Biol Chem 276: 45921–45927. https://doi.org/10.1074/jbc.M107726200
Takano A, Isojima Y, Nagai K (2004) Identification of mPer1 phosphorylation sites responsible for the nuclear entry. J Biol Chem 279: 32578–32585. https://doi.org/10.1074/jbc.M403433200
Aryal RP, Kwak PB, Tamayo AG, Gebert M, Chiu PL, Walz T, Weitz CJ (2017) Macromolecular Assemblies of the Mammalian Circadian Clock. Mol Cell 67: 770–782. https://doi.org/10.1016/j.molcel.2017.07.017
Eng GWL, Edison, Virshup DM (2017) Site-specific phosphorylation of casein kinase 1 δ (CK1δ) regulates its activity towards the circadian regulator PER2. PLoS One 12. https://doi.org/10.1371/journal.pone.0177834
Paul JR, McKeown AS, Davis JA, Totsch SK, Mintz EM, Kraft TW, Cowell RM, Gamble KL (2017) Glycogen synthase kinase 3 regulates photic signaling in the suprachiasmatic nucleus. Eur J Neurosci 45: 1102–1110. https://doi.org/10.1111/ejn.13549
Leloup J-C, Goldbeter A (2011) Modelling the dual role of Per phosphorylation and its effect on the period and phase of the mammalian circadian clock. IET Syst Biol 5: 44–49. https://doi.org/10.1049/iet-syb.2009.0068
Yin L, Wang J, Klein PS, Lazar MA (2006) Nuclear Receptor Rev-erbα Is a Critical Lithium-Sensitive Component of the Circadian Clock. Science (80) 311: 1002–1005. https://doi.org/10.1126/science.1121613
Kurabayashi N, Hirota T, Sakai M, Sanada K, Fukada Y (2010) DYRK1A and Glycogen Synthase Kinase 3β, a Dual-Kinase Mechanism Directing Proteasomal Degradation of CRY2 for Circadian Timekeeping. Mol Cell Biol 30: 1757–1768. https://doi.org/10.1128/MCB.01047-09
Sahar S, Zocchi L, Kinoshita C, Borrelli E, Sassone-Corsi P (2010) Regulation of BMAL1 Protein Stability and Circadian Function by GSK3β-Mediated Phosphorylation. PLoS One 5: e8561. https://doi.org/10.1371/journal.pone.0008561
Spengler ML, Kuropatwinski KK, Schumer M, Antoch M (2009) A serine cluster mediates BMAL1-dependent CLOCK phosphorylation and degradation. Cell Cycle 8: 4138–4146. https://doi.org/10.4161/cc.8.24.10273
Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U (2008) SIRT1 Regulates Circadian Clock Gene Expression through PER2 Deacetylation. Cell 134: 317–328. https://doi.org/10.1016/j.cell.2008.06.050
Wang R-H, Zhao T, Cui K, Hu G, Chen Q, Chen W, Wang X-W, Soto-Gutierrez A, Zhao K, Deng C-X (2016) Negative reciprocal regulation between Sirt1 and Per2 modulates the circadian clock and aging. Sci Rep 6: 28633. https://doi.org/10.1038/srep28633
Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P (2008) The NAD+-Dependent Deacetylase SIRT1 Modulates CLOCK-Mediated Chromatin Remodeling and Circadian Control. Cell 134: 329–340. https://doi.org/10.1016/j.cell.2008.07.002
Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P (2009) Circadian Control of the NAD + Salvage Pathway by CLOCK-SIRT1. Science (80)324: 654–657. https://doi.org/10.1126/science.1170803
Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong H-K, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J (2009) Circadian Clock Feedback Cycle Through NAMPT-Mediated NAD+ Biosynthesis. Science (80) 324: 651–654. https://doi.org/10.1126/science.1171641
Chang H-C, Guarente L (2013) SIRT1 Mediates Central Circadian Control in the SCN by a Mechanism that Decays with Aging. Cell 153: 1448–1460. https://doi.org/10.1016/j.cell.2013.05.027
Orozco-Solis R, Ramadori G, Coppari R, Sassone-Corsi P (2015) SIRT1 relays nutritional inputs to the circadian clock through the Sf1 neurons of the ventromedial hypothalamus. Endocrinology (United States) 156: 2174–2184. https://doi.org/10.1210/en.2014-1805
Schibler U (2021) BMAL1 dephosphorylation determines the pace of the circadian clock. Genes Dev 35: 1076–1078. https://doi.org/10.1101/gad.348801.121
DiTacchio L, Le HD, Vollmers C, Hatori M, Witcher M, Secombe J, Panda S (2011) Histone Lysine Demethylase JARID1a Activates CLOCK-BMAL1 and Influences the Circadian Clock. Science (80) 333: 1881–1885.https://doi.org/10.1126/science.1206022
Gareau JR, Lima CD (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11: 861–871. https://doi.org/10.1038/nrm3011
Chen L-C, Hsieh Y-L, Tan GYT, Kuo T-Y, Chou Y-C, Hsu P-H, Hwang-Verslues WW (2021) Differential effects of SUMO1 and SUMO2 on circadian protein PER2 stability and function. Sci Rep 11: 14431. https://doi.org/10.1038/s41598-021-93933-y
Fernandez DC, Fogerson PM, Lazzerini Ospri L, Thomsen MB, Layne RM, Severin D, Zhan J, Singer JH, Kirkwood A, Zhao H, Berson DM, Hattar S (2018) Light Affects Mood and Learning through Distinct Retina-Brain Pathways. Cell 175: 71–84. https://doi.org/10.1016/j.cell.2018.08.004
Falchi F, Cinzano P, Duriscoe D, Kyba CCM, Elvidge CD, Baugh K, Portnov BA, Rybnikova NA, Furgoni R (2016) The new world atlas of artificial night sky brightness. Sci Adv 2. https://doi.org/10.1126/sciadv.1600377
Tähkämö L, Partonen T, Pesonen A-K (2019) Systematic review of light exposure impact on human circadian rhythm. Chronobiol Int 36: 151–170. https://doi.org/10.1080/07420528.2018.1527773
Faulkner SM, Bee PE, Meyer N, Dijk D-J, Drake RJ (2019) Light therapies to improve sleep in intrinsic circadian rhythm sleep disorders and neuro-psychiatric illness: A systematic review and meta-analysis. Sleep Med Rev 46: 108–123. https://doi.org/10.1016/j.smrv.2019.04.012
Al-Karawi D, Jubair L (2016) Bright light therapy for nonseasonal depression: Meta-analysis of clinical trials. J Affect Disord 198: 64–71. https://doi.org/10.1016/j.jad.2016.03.016
Zee PC, Goldstein CA (2010) Treatment of Shift Work Disorder and Jet Lag. Curr Treat Options Neurol 12: 396–411. https://doi.org/10.1007/s11940-010-0090-9
Rundle AG, Revenson TA, Friedman M (2018) Business Travel and Behavioral and Mental Health. J Occup Environ Med 60: 612–616. https://doi.org/10.1097/JOM.0000000000001262
Li Y, Androulakis IP (2021) Light entrainment of the SCN circadian clock and implications for personalized alterations of corticosterone rhythms in shift work and jet lag. Sci Rep 11: 17929. https://doi.org/10.1038/s41598-021-97019-7
Castilhos Beauvalet J, Luísa Quiles C, Alves Braga de Oliveira M, Vieira Ilgenfritz CA, Hidalgo MP, Comiran Tonon A (2017) Social jetlag in health and behavioral research: a systematic review. Chron Physiol Ther 7: 19–31. https://doi.org/10.2147/CPT.S108750
Knapen SE, Riemersma-van der Lek RF, Antypa N, Meesters Y, Penninx BWJH, Schoevers RA (2018) Social jetlag and depression status: Results obtained from the Netherlands Study of Depression and Anxiety. Chronobiol Int 35: 1–7. https://doi.org/10.1080/07420528.2017.1374966
Benedetti F, Riccaboni R, Dallaspezia S, Locatelli C, Smeraldi E, Colombo C (2015) Effects of CLOCK gene variants and early stress on hopelessness and suicide in bipolar depression. Chronobiol Int 32: 1156–1161. https://doi.org/10.3109/07420528.2015.1060603
Rybakowski JK, Dmitrzak-Weglarz M, Dembinska-Krajewska D, Hauser J, Akiskal KK, Akiskal HH (2014) Polymorphism of circadian clock genes and temperamental dimensions of the TEMPS-A in bipolar disorder. J Affect Disord 159: 80–84. https://doi.org/10.1016/j.jad.2014.02.024
Jankowski KS, Dmitrzak-Weglarz M (2017) ARNTL, CLOCK and PER3 polymorphisms – links with chronotype and affective dimensions. Chronobiol Int 34: 1105–1113. https://doi.org/10.1080/07420528.2017.1343341
Kim H-I, Lee H-J, Cho C-H, Kang S-G, Yoon H-K, Park Y-M, Lee S-H, Moon J-H, Song H-M, Lee E, Kim L (2015) Association of CLOCK, ARNTL, and NPAS2 gene polymorphisms and seasonal variations in mood and behavior. Chronobiol Int 32: 785–791. https://doi.org/10.3109/07420528.2015.1049613
Liberman AR, Halitjaha L, Ay A, Ingram KK (2018) Modeling Strengthens Molecular Link between Circadian Polymorphisms and Major Mood Disorders. J Biol Rhythms 33: 318–336. https://doi.org/10.1177/0748730418764540
Melhuish Beaupre L, Brown GM, Kennedy JL (2020) Circadian genes in major depressive disorder. World J Biol Psychiatry 21: 80–90. https://doi.org/10.1080/15622975.2018.1500028
Hua P, Liu W, Chen D, Zhao Y, Chen L, Zhang N, Wang C, Guo S, Wang L, Xiao H, Kuo S-H (2014) Cry1 and Tef gene polymorphisms are associated with major depressive disorder in the Chinese population. J Affect Disord 157: 100–103. https://doi.org/10.1016/j.jad.2013.11.019
Drago A, Monti B, De Ronchi D, Serretti A (2015) CRY1 Variations Impacts on the Depressive Relapse Rate in a Sample of Bipolar Patients. Psychiatr Invest 12: 118. https://doi.org/10.4306/pi.2015.12.1.118
D’Souza T, Rajkumar AP (2020) Systematic review of genetic variants associated with cognitive impairment and depressive symptoms in Parkinson’s disease. Acta Neuropsychiatr 32: 10–22. https://doi.org/10.1017/neu.2019.28
Fiedorowicz JG, Coryell WH, Akhter A, Ellingrod VL (2012) Cryptochrome 2 variants, chronicity, and seasonality of mood disorders. Psychiatr Genet 22: 305–306. https://doi.org/10.1097/YPG.0b013e3283539594
Kovanen L, Kaunisto M, Donner K, Saarikoski ST, Partonen T (2013) CRY2 Genetic Variants Associate with Dysthymia. PLoS One 8: e71450. https://doi.org/10.1371/journal.pone.0071450
Kovanen L, Donner K, Kaunisto M, Partonen T (2017) PRKCDBP (CAVIN3) and CRY2 associate with major depressive disorder. J Affect Disord 207: 136–140. https://doi.org/10.1016/j.jad.2016.09.034
Parekh PK, Becker-Krail D, Sundaravelu P, Ishigaki S, Okado H, Sobue G, Huang Y, McClung CA (2018) Altered GluA1 (Gria1) Function and Accumbal Synaptic Plasticity in the ClockΔ19 Model of Bipolar Mania. Biol Psychiatry 84: 817–826. https://doi.org/10.1016/j.biopsych.2017.06.022
Kozikowski AP, Gunosewoyo H, Guo S, Gaisina IN, Walter RL, Ketcherside A, McClung CA, Mesecar AD, Caldarone B (2011) Identification of a Glycogen Synthase Kinase-3β Inhibitor that Attenuates Hyperactivity in CLOCK Mutant Mice. Chem Med Chem 6: 1593–1602. https://doi.org/10.1002/cmdc.201100188
Arey R, McClung CA (2012) An inhibitor of casein kinase 1 ε/δ partially normalizes the manic-like behaviors of the ClockΔ19 mouse. Behav Pharmacol 23: 392–396. https://doi.org/10.1097/FBP.0b013e32835651fd
Mukherjee S, Coque L, Cao J-L, Kumar J, Chakravarty S, Asaithamby A, Graham A, Gordon E, Enwright JF, DiLeone RJ, Birnbaum SG, Cooper DC, McClung CA (2010) Knockdown of Clock in the Ventral Tegmental Area Through RNA Interference Results in a Mixed State of Mania and Depression-Like Behavior. Biol Psychiatry 68: 503–511. https://doi.org/10.1016/j.biopsych.2010.04.031
Qiu P, Jiang J, Liu Z, Cai Y, Huang T, Wang Y, Liu Q, Nie Y, Liu F, Cheng J, Li Q, Tang Y-C, Poo M, Sun Q, Chang H-C (2019) BMAL1 knockout macaque monkeys display reduced sleep and psychiatric disorders. Natl Sci Rev 6: 87–100. https://doi.org/10.1093/nsr/nwz002
Landgraf D, Long JE, Proulx CD, Barandas R, Malinow R, Welsh DK (2016) Genetic Disruption of Circadian Rhythms in the Suprachiasmatic Nucleus Causes Helplessness, Behavioral Despair, and Anxiety-like Behavior in Mice. Biol Psychiatry 80: 827–835. https://doi.org/10.1016/j.biopsych.2016.03.1050
Leliavski A, Shostak A, Husse J, Oster H (2014) Impaired Glucocorticoid Production and Response to Stress in Arntl-Deficient Male Mice. Endocrinology 155: 133–142. https://doi.org/10.1210/en.2013-1531
Jager J, O’Brien WT, Manlove J, Krizman EN, Fang B, Gerhart-Hines Z, Robinson MB, Klein PS, Lazar MA (2014) Behavioral Changes and Dopaminergic Dysregulation in Mice Lacking the Nuclear Receptor Rev-erbα. Mol Endocrinol 28: 490–498. https://doi.org/10.1210/me.2013-1351
Spencer S, Falcon E, Kumar J, Krishnan V, Mukherjee S, Birnbaum SG, McClung CA (2013) Circadian genes Period 1 and Period 2 in the nucleus accumbens regulate anxiety-related behavior. Eur J Neurosci 37: 242–250. https://doi.org/10.1111/ejn.12010
Li Y, Li G, Li J, Cai X, Sun Y, Zhang B, Zhao H (2021) Depression-like behavior is associated with lower Per2 mRNA expression in the lateral habenula of rats. Genes, Brain Behav 20. https://doi.org/10.1111/gbb.12702
Zhang L, Hirano A, Hsu P-K, Jones CR, Sakai N, Okuro M, McMahon T, Yamazaki M, Xu Y, Saigoh N, Saigoh K, Lin S-T, Kaasik K, Nishino S, Ptáček LJ, Fu Y-H (2016) A PERIOD3 variant causes a circadian phenotype and is associated with a seasonal mood trait. Proc Natl Acad Sci U S A 113. https://doi.org/10.1073/pnas.1600039113
Huang J, Zhong Z, Wang M, Chen X, Tan Y, Zhang S, He W, He X, Huang G, Lu H, Wu P, Che Y, Yan Y-L, Postlethwait JH, Chen W, Wang H (2015) Circadian Modulation of Dopamine Levels and Dopaminergic Neuron Development Contributes to Attention Deficiency and Hyperactive Behavior. J Neurosci 35: 2572–2587. https://doi.org/10.1523/JNEUROSCI.2551-14.2015
De Bundel D, Gangarossa G, Biever A, Bonnefont X, Valjent E (2013) Cognitive dysfunction, elevated anxiety, and reduced cocaine response in circadian clock-deficient cryptochrome knockout mice. Front Behav Neurosci 7. https://doi.org/10.3389/fnbeh.2013.00152
Tapia-Osorio A, Salgado-Delgado R, Angeles-Castellanos M, Escobar C (2013) Disruption of circadian rhythms due to chronic constant light leads to depressive and anxiety-like behaviors in the rat. Behav Brain Res 252: 1–9. https://doi.org/10.1016/j.bbr.2013.05.028
Walker WH, Borniger JC, Gaudier-Diaz MM, Hecmarie Meléndez-Fernández O, Pascoe JL, Courtney DeVries A, Nelson RJ (2020) Acute exposure to low-level light at night is sufficient to induce neurological changes and depressive-like behavior. Mol Psychiatry 25: 1080–1093. https://doi.org/10.1038/s41380-019-0430-4
Borniger JC, McHenry ZD, Abi Salloum BA, Nelson RJ (2014) Exposure to dim light at night during early development increases adult anxiety-like responses. Physiol Behav 133: 99–106. https://doi.org/10.1016/j.physbeh.2014.05.012
Bedrosian TA, Galan A, Vaughn CA, Weil ZM, Nelson RJ (2013) Light at Night Alters Daily Patterns of Cortisol and Clock Proteins in Female Siberian Hamsters. J Neuroendocrinol 25: 590–596. https://doi.org/10.1111/jne.12036
Karatsoreos IN, Bhagat S, Bloss EB, Morrison JH, McEwen BS (2011) Disruption of circadian clocks has ramifications for metabolism, brain, and behavior. Proc Natl Acad Sci U S A 108: 1657–1662. https://doi.org/10.1073/pnas.1018375108
Valvassori SS, Resende WR, Dal-Pont G, Sangaletti-Pereira H, Gava FF, Peterle BR, Carvalho AF, Varela RB, Dal-Pizzol F, Quevedo J (2017) Lithium ameliorates sleep deprivation-induced mania-like behavior, hypothalamic-pituitary-adrenal (HPA) axis alterations, oxidative stress and elevations of cytokine concentrations in the brain and serum of mice. Bipolar Disord 19: 246–258. https://doi.org/10.1111/bdi.12503
Wells AM, Ridener E, Bourbonais CA, Kim W, Pantazopoulos H, Carroll FI, Kim K-S, Cohen BM, Carlezon WA (2017) Effects of Chronic Social Defeat Stress on Sleep and Circadian Rhythms Are Mitigated by Kappa-Opioid Receptor Antagonism. J Neurosci 37: 7656–7668.https://doi.org/10.1523/JNEUROSCI.0885-17.2017
Henderson F, Vialou V, El Mestikawy S, Fabre V (2017) Effects of Social Defeat Stress on Sleep in Mice. Front Behav Neurosci 28(11): 227. https://doi.org/10.3389/fnbeh.2017.00227
Ota SM, Suchecki D, Meerlo P (2018) Chronic social defeat stress suppresses locomotor activity but does not affect the free-running circadian period of the activity rhythm in mice. Neurobiol Sleep Circadian Rhythm 5: 1–7. https://doi.org/10.1016/j.nbscr.2018.03.002
Olejníková L, Polidarová L, Sumová A (2018) Stress affects expression of the clock gene Bmal1 in the suprachiasmatic nucleus of neonatal rats via glucocorticoid-dependent mechanism. Acta Physiol 223: e13020. https://doi.org/10.1111/apha.13020
Levitas-Djerbi T, Appelbaum L (2017) Modeling sleep and neuropsychiatric disorders in zebrafish. Curr Opin Neurobiol 44: 89–93. https://doi.org/10.1016/j.conb.2017.02.017
Tang Y-Q, Li Z-R, Zhang S-Z, Mi P, Chen D-Y, Feng X-Z (2019) Venlafaxine plus melatonin ameliorate reserpine-induced depression-like behavior in zebrafish. Neurotoxicol Teratol 76: 106835. https://doi.org/10.1016/j.ntt.2019.106835
Дополнительные материалы отсутствуют.
Инструменты
Российский физиологический журнал им. И.М. Сеченова


