Российский физиологический журнал им. И.М. Сеченова, 2023, T. 109, № 7, стр. 819-843
GCaMP – семейство однофлуорофорных генетически-кодируемых кальциевых индикаторов
А. И. Ерофеев 1, *, Е. К. Винокуров 1, О. Л. Власова 1, И. Б. Безпрозванный 1, 2
1 Лаборатория молекулярной нейродегенерации, Институт биомедицинских систем
и биотехнологий, Санкт-Петербургский Политехнический университет Петра Великого
Санкт-Петербург, Россия
2 Отделение физиологии, Юго-Западный медицинский центр Техасского университета,
Даллас
Техас, США
* E-mail: alexandr.erofeew@gmail.com
Поступила в редакцию 04.05.2023
После доработки 16.06.2023
Принята к публикации 17.06.2023
- EDN: XHKZHO
- DOI: 10.31857/S0869813923070038
Аннотация
Однофлуорофорные генетически кодируемые кальциевые индикаторы, такие как GCaMP, являются широко используемыми инструментами для изучения нейрональной активности. Их главным преимуществом является способность в режиме реального времени и с высокой чувствительностью реагировать на изменения концентрации ионов кальция в клетке. Это особенно важно для исследования нейрональных процессов и нейронных ансамблей, где кальциевые сигналы играют ключевую роль в передаче информации. Данный обзор рассматривает семейство кальциевых индикаторов GCaMP, описывает их разновидности, особенности и перспективы использования для визуализации нейронной активности. Особое внимание уделяется тенденции совершенствования GCaMP, в частности направленной на расширение их спектральных свойств и способности регистрировать высокочастотную спайковую активность.
СОКРАЩЕНИЯ И ТЕРМИНЫ
Апокальмодулин (apoCaM) – свободная от Ca2+ форма кальмодулина, которая может взаимодействовать с определенными белками в отсутствие кальция.
Биолюминесценция – это естественное явление, которое возникает в результате биохимической реакции, включающей окисление небольшой молекулы люциферина, внутри живых организмов.
ГеККИ (генетически-кодируемые кальциевые индикаторы) – тип биосенсоров, используемых для обнаружения и измерения концентрации ионов кальция в живых клетках.
Деплеция – процесс уменьшения или исчерпания какого-либо вещества в клетке.
Кальциевый транзиент – это изменение концентрации ионов кальция внутри клетки в ответ на определенный сигнал, например, сигнал от нейромедиатора или гормона.
Кальмодулин – это небольшой кальций-связывающий белок, который участвует во многих клеточных процессах, включая сокращение мышц, высвобождение нейротрансмиттеров и транскрипцию генов. Когда кальций связывается с кальмодулином, он претерпевает конформационные изменения, что позволяет ему взаимодействовать с различными целевыми белками, включая ферменты и ионные каналы.
Квантовый выход (Φ) – это показатель эффективности преобразования поглощенного света в флуоресценцию, представляет собой отношение числа испускаемых фотонов (флуоресцентный сигнал) к числу поглощенных фотонов (поглощенный свет). Квантовый выход измеряется в процентах или в долях единицы (от 0 до 1).
Константа диссоциации по кислотности (pKa) – это значение pH, при котором половина молекул индикатора GCaMP находится в протонированной форме (несвязанной с кальцием) и половина в депротонированной форме (связанной с кальцием).
Коэффициент экстинкции (ε, молярный коэффициент экстинкции) – это мера, которая характеризует способность вещества поглощать свет определенной длины волны, определяется как отношение поглощенной интенсивности света к концентрации GCaMP в растворе. Коэффициент экстинкции позволяет оценить, насколько эффективно GCaMP поглощает свет определенной длины волны, что влияет на его способность обнаруживать изменения концентрации кальция в клетке. Коэффициент экстинкции измеряется в литрах, деленных на моль и на сантиметр (л моль−1 см−1) или в молярных единицах экстинкции (M−1 см−1).
Мотив – участок белка, который имеет определенную структуру и выполняет определенную функцию.
Нейриты (или аксоны) – это длинные, тонкие ветви нейрона, которые передают информацию в виде электрических импульсов от тела клетки (сомы) к другим нейронам, мышцам или железам.
Нейронный ансамбль – это группа нейронов, которые активируются синхронно и совместно участвуют в выполнении определенных функций и задач в мозге.
Нейропиль – это область нервной ткани, где нейроны устанавливают связи между собой и другими клетками.
Опсины – это белки, которые содержатся в глазах живых организмов и участвуют в процессе зрения. Они связываются с пигментами, такими как ретиналь, и образуют фотоприемники, которые способны реагировать на световые стимулы и транслировать их в нервные импульсы для дальнейшей обработки в головном мозге.
Оптическая электрофизиология (Оптофизиология) – это область научных исследований, которая объединяет методы оптики и электрофизиологии для изучения электрической активности клеток и тканей в организмах. Она использует оптические методы, такие как флуоресценция, для измерения электрических потенциалов и токов в нервной системе, мышечных тканях и других типах тканей, которые генерируют электрическую активность.
Пептид M13 – это последовательность из 13 аминокислот, которая обладает высоким сродством к кальмодулину и может специфически связываться с ним даже в присутствии других кальций-связывающих белков.
Пресинаптическая терминаль (бутон) – расширение конца нервного волокна, которое содержит мембранные белки (синаптические белки), необходимые для передачи нервного импульса на постсинаптический нейрон или эффекторную клетку.
Спайк – короткий электрический импульс, который генерируется нейроном и передается по аксону к другим нейронам или эффекторным клеткам, таким как мышцы или железы. Спайк часто используется для описания электрической активности нейронов на уровне целых нейронных сетей или организмов, в отличие от термина “потенциал действия”, который обычно используется для описания физиологических процессов, происходящих внутри нейрона.
Тропизм вируса – это способность вируса инфицировать определенные типы клеток или тканей в организме своего хозяина.
Тропонин-С – это белковый компонент тропонинового комплекса, состоящий из двух глобулярных доменов, каждый из которых связывает по два иона кальция, изменяя конформацию комплекса и вызывая сокращение мышечной клетки. Тропонин-С широко используется в диагностике острого коронарного синдрома и других сердечно-сосудистых заболеваний, так как его уровень в крови может указывать на повреждение миокарда.
ФБ – флуоресцентный белок.
Флуоресценция – это явление, при котором вещество поглощает свет определенной длины волны, а затем испускает свет большей длины волны, обычно с меньшей энергией.
Флуорофор – это молекула, которая поглощает свет определенной длины волны, а затем излучает свет большей длины волны. Флуорофоры могут быть естественного происхождения, например, зеленый флуоресцентный белок (GFP), обнаруженный в медузах, или синтетические, такие как флуоресцеин или родамин.
Цитоморбидность – нарушение функции клеток или их структуры, вызванное различными факторами, такими как инфекции, токсины, нарушения обмена веществ и другие.
Цитотоксичность – это способность вещества или микроорганизма повреждать или уничтожать клетки. Например, повышенный уровень кальция в нейроне может вызвать активацию кальций-зависимых протеаз, таких как кальпаин и катепсин, которые могут разрушать клеточные белки, включая белки клеточного скелета.
Шипик (спайклет) – это маленький выступ на дендритах или соме нейрона, который может контактировать с окружающими клетками, в том числе и другими нейронами. Шипики обычно играют важную роль в формировании и поддержании синаптических контактов между нейронами.
Aequorin – это фотопротеин, выделенный из медузы Aequorea victoria, кальций-чувствительный белок, который излучает синий свет, когда связывается с ионами кальция.
Arch (Archaerhodopsin) – архродопсин, семейство ретиналь-содержащих фоторецепторов, обнаруженных у одноклеточных микроорганизмов Halobacterium и Halorubrum.
CaMKK (Calcium/calmodulin-dependent protein kinase kinase) – киназа кальций/кальмодулин-зависимой протеинкиназы.
ChR2 (Channelrhodopsin-2) – каналородопсин-2, семейство ретиналь-содержащих фоторецепторов, обнаруженных у одноклеточных синих водорослей Chlamydomonas reinhardtii.
ckkap (CaM-dependent kinase kinase peptide) – пептид CaM-зависимой киназы.
cpGFP (circularly permuted green fluorescent protein) – круговой пермутированный зеленый флуоресцентный белок. Вариант GFP, в котором аминокислотные последовательности были переставлены для создания круговой формы белка. Обладает более высокой интенсивностью флуоресценции и более коротким временем перехода между состояниями, что делает его более подходящим для регистрации быстрых процессов в клетках.
Cre-рекомбиназа – это фермент, который используется в генетической инженерии для управления экспрессией генов. Cre-рекомбиназа может распознавать специфические ДНК-последовательности, называемые “локусами”, и вызывать рекомбинацию между ними. Это позволяет исследователям удалять, вставлять или переставлять определенные гены в геноме организма.
EGFP (Enhanced Green Fluorescent Protein) – улучшенный вариант GFP, который имеет более яркую и стабильную флуоресценцию. EGFP менее склонен к деградации, т.е. к потере флуоресценции с течением времени.
EGTA (Этиленгликоль-бис(2-аминоэтилэфир)-N,N,N',N'-тетрауксусная кислота) – органическое соединение, используемое в биохимии и молекулярной биологии в качестве хелатирующего агента. EGTA широко применяется для связывания и удаления дивалентных ионов металлов, особенно ионов кальция (Ca2+), из растворов.
eNOS (Endothelial Nitric Oxide Synthase) – эндотелиальная синтаза оксида азота.
GFP – зеленый флуоресцентный белок, впервые обнаружен в медузе Aequorea victoria в 1962 г.
HR (Halorhodopsin) – галородопсин, семейство ретиналь-содержащих фоторецепторов, обнаруженных у одноклеточных микроорганизмов, известных как галобактерии.
mApple – один из видов мутантного зеленого флуоресцентного белка, имеющий красновато-оранжевый цвет, более высокую стабильность и яркость, чем оригинальный GFP.
MOPS – это химическое соединение, используемое в биохимических и биологических исследованиях в качестве буфера. Он является слабой кислотой с pKa около 7.2, что делает его эффективным для поддержания постоянного pH в диапазоне 6.5–7.9.
mRuby – один из видов мутантного зеленого флуоресцентного белка (GFP), имеющий красный цвет, более высокую стабильность и яркость, чем оригинальный GFP.
RFP (Red Fluorescent Protein) – это красный флуоресцентный белок, который используется в биологических исследованиях для маркировки клеток и белков.
SNR (Signal-to-Noise Ratio, соотношение сигнал–шум) – это показатель, характеризующий соотношение между уровнем сигнала и уровнем шума в измерении или эксперименте.
ВВЕДЕНИЕ
Визуализация кальция (Ca2+), которая представляет собой оптическое измерение концентрации кальция с помощью специальных флуоресцентных индикаторов [1, 2], имеет долгую и богатую историю, восходящую к концу 1960-х – началу 1970-х годов. В то время исследователи впервые обнаружили, что изменения уровня внутриклеточного кальция связаны с активностью нейронов [3, 4], и начали использовать синтетические флуоресцентные кальций-чувствительные красители (например, Quin-2, Fura-2 или Fluo-3) в качестве индикаторов активности нейронов [5–7]. Хотя это была революционная технология, необходимость доставки этих молекул с помощью патч-пипетки или объемной внеклеточной загрузки в липофильных условиях ограничивала специфические для конкретного типа клеток условия доставки и визуализации [8]. В 1980-х и 1990-х годах достижения в области микроскопии и технологий визуализации, включая изобретение электронного микроскопа, развитие флуоресцентной микроскопии, изобретение усовершенствованных стереомикроскопов, развитие оптических и спектральных методов количественной визуализации, развитие методов маркировки флуоресцентных белков, позволили исследователям визуализировать изменения внутриклеточного уровня кальция в нейронах как in vitro, так и in vivo в режиме реального времени [9–11]. Впоследствии область визуализации кальция в нейробиологии продолжала развиваться и расширяться, что было обусловлено технологическим прогрессом и растущей потребностью в более сложных методах регистрации активности нейронов. Это привело к разработке новых кальциевых индикаторов, таких как генетически-кодируемые кальциевые индикаторы (ГеККИ), которые позволили регистрировать изменения уровня внутриклеточного кальция с высокой чувствительностью и специфичностью [12, 13]. Сегодня кальциевая визуализация широко используется в исследованиях для изучения синаптической передачи, регистрации активности нейронов и понимания влияния лекарств и заболеваний на мозг. Данный метод остается важнейшим инструментом для исследователей в этой области и продолжает играть важную роль в изучении нервной системы.
ГЕНЕТИЧЕСКИ-КОДИРУЕМЫЕ ИНДИКАТОРЫ КАЛЬЦИЯ
Ион кальция (Ca2+) является распространенным вторичным посредником в клеточной передаче сигналов, с помощью которого регулируются многие биологические явления, в частности нейронная активность [3, 4, 14–16]. Уровень цитоплазматического кальция в состоянии покоя обычно составляет 50–100 нМ, однако активность нейронов стимулирует его приток [1, 13]. Во время распространения потенциала действия (ПД), потенциал-зависимые кальциевые каналы приводят к увеличению соматического кальция до ~1 мкМ [17, 18]. Поэтому ПД могут быть косвенно оценены путем измерения изменений внутриклеточного Ca2+ [1, 19–21]. Для получения более подробной информации о различных формах возбуждения Ca2+, о том, как они различаются в разных типах нейронов, и о биохимических механизмах связывания Ca2+ с флуоресцентными изменениями читатели могу ознакомиться в обзорах [1, 13, 22].
Первые эксперименты по визуализации внутриклеточных изменений Ca2+ [23, 24] были проведены с использованием Ca2+-чувствительного биолюминесцентного белка aequorin [25]. Эти эксперименты предшествовали появлению генетически-кодированных Ca2+-индикаторов [26], так же, как открытие зеленого флуоресцентного белка (GFP) [25], создание различных вариантов GFP [27] и биохимическое изучение связывания Ca2+ путем слияния кальмодулина с его связывающим пептидом M13, полученным из киназы легкой цепи миозина [28]. Эра современных ГеККИ началась с появлением биосенсоров на основе флуоресцентных белков (ФБ), способных определять изменения уровня внутриклеточного кальция [21, 29], напряжения мембраны [30] и секреции синаптических пузырьков [31].
Генетически-кодируемые кальциевые индикаторы – это тип биосенсоров, используемых для измерения концентрации ионов кальция в живых клетках. ГеККИ создаются путем слияния кальций-связывающего белка, такого как кальмодулин или тропонин-С [26, 32], с флуоресцентным белком, например, GFP, для регистрации внутриклеточного уровня кальция. Существует несколько типов ГеККИ, включая однофлуорофорные ГеККИ (интенсиометрические), двухфлуорофорные ГеККИ (резонансного переноса энергии Фёрстера, FRET, ратиометрические) [33–35] и ГеККИ на основе флуоресцентных белков, отличных от GFP. ГеККИ наиболее широко используются для прижизненной визуализации в модельных системах, включая червей [36, 37], зебрафиш [38], мух [39], грызунов [40, 41] и нечеловеческих приматов [42, 43].
В данном обзоре речь пойдет об интенсиометрических Ca2+-сенсорах, таких как GCaMP, т.к. этот тип ГеККИ является одним из самых распространенных сенсоров для визуализации нейронной активности [44]. Данный обзор будет полезен читателям, начинающим работать в области флуоресцентной визуализации нейронной активности.
ПРЕИМУЩЕСТВА И НЕДОСТАТКИ ГЕНЕТИЧЕСКИ-КОДИРУЕМЫХ КАЛЬЦИЕВЫХ ИНДИКАТОРОВ
Преимущество ГеККИ в том, что они позволяют проводить хроническую неинвазивную визуализацию определенных клеток и компартментов [45] за счет оптической флуоресцентной визуализации, которая в свою очередь позволяет одновременно исследовать большие популяции нейронов, избежать серьезного повреждения интересующих клеток [13]. ГеККИ могут быть введены с помощью вирусной доставки генов или трансгенеза (постоянной геномной модификации). Генетический контроль снижает фоновый сигнал от источников, не являющихся объектом исследования, повышая соотношение сигнал–шум (SNR) [46] и позволяя исследовать популяцию клеток, которая анатомически или функционально связана. Например, ГеККИ позволяют специфически маркировать нейроны прямых и непрямых путей в стриатуме [47], а также возбуждающие [48] и тормозные [49] нейроны. Также ГеККИ позволяют одновременно регистрировать данные с большой популяции клеток с лучшим пространственным разрешением, чем электрофизиологические методы [50, 51]. Это не означает, что оптическая визуализация устраняет необходимость применения электрических методов в нейробиологии. Напротив, методы электрической регистрации также прогрессируют и могут предложить лучшее или сопоставимое пространственное разрешение [52].
Возможность проводить хроническую регистрацию одних и тех же клеток в течение длительного периода времени [34, 41, 53] является одним из ключевых преимуществ ГеККИ. Их применение дало возможность получать изображения динамики уровней внеклеточных нейротрансмиттеров и нейромодуляторов у свободно передвигающихся животных [54, 55]. Кроме того, важным преимуществом визуализации Ca2+ является совместимость этого метода с другими оптическими методами. Например, сочетание визуализации Ca2+ с оптогенетическими подходами позволяет реализовать оптическую электрофизиологию, в которой одновременно происходит визуализация и модуляция светом одних и тех же групп нейронов [56].
К недостаткам ГеККИ можно отнести необходимость подбора уровня экспрессии. Низкий уровень экспрессии исключает визуализацию сенсора и снижает SNR. Он также требует визуализации с более высокой мощностью источника света и более длительным временем экспозиции, что может привести к фототоксичности, снижению скорости визуализации и увеличению фотообесцвечивания. С другой стороны, высокий уровень экспрессии подразумевает использование меньшей мощности источника возбуждения, но также увеличивает буферизацию кальция и может нарушить сигнальные пути клетки, что может привести к цитотоксичности и цитоморбидности [12, 41, 57, 58]. Проблема, как правило, решается выбором соответствующего задачам исследования серотипа вируса с различным тропизмом для разных типов клеток и эмпирическим подбором его концентрации [59, 60]. Среди новых серотипов с улучшенной трансдукцией и новыми свойствами доставки следует отметить AAV-PHP.eB. Данный серотип является производным AAV9, который эффективно преодолевает гематоэнцефалический барьер после внутривенной инъекции [61], обеспечивая удобный способ экспрессии необходимых индикаторов в нескольких областях мозга грызунов [62]. Индикаторы с высоким сродством к Ca2+ могут нарушать собственные Ca2+-зависимые процессы в клетке [63, 64]. Так, например, варианты ГеККИ на основе кальмодулина могут влиять на работу кальциевых каналов L-типа (CaV1), препятствуя как их открытию, так и передаче сигнала [65]. Решением данной проблемы может быть использование бескальциевой формы кальмодулина (apoCaM) [66]. Кроме того, ГеККИ могут связываться с кальцием не только в местах, где его концентрация повышена, но и там, где кальция нет, но есть другие катионы, например, магний. Это приводит к ложноположительным сигналам, которые могут искажать интерпретацию данных [67]. Одним из вариантов устранения проблемы высокого сродства ГеККИ к кальцию является создание новых версий индикатора с измененными свойствами [68]. Например, существуют индикаторы с низким сродством к магнию, такие как GCaMP6, которые позволяют снизить количество ложноположительных сигналов [69]. Также возможно использование индикаторов, которые реагируют на другие ионы, например, натрий [70] или калий [71]. ГеККИ, такие как GCaMP, имеют медленную (миллисекундную) кинетику, что может затруднять наблюдение быстро изменяющейся активности клеток, т.е. регистрацию высокочастотных потенциалов действия [72, 73].
В целом, несмотря на проблемы, ГеККИ в сочетании с современными методами микроскопии сегодня широко используются для исследования нейронной активности в самых разных условиях как in vitro, так и in vivo [74].
ОДНОФЛУОРОФОРНЫЕ ГЕККИ
Первым однофлуорофорным (интенсиометрическим) ГеККИ был Camgaroo – индикатор кальция, реализованный на основе кальмодулина (CaM) и желтого флуоресцентного белка (YFP) [75]. Camgaroo был первым индикатором, в котором Ca2+-зависимые конформационные изменения в CaM, вызывали ионизацию хромофора, которая приводила к увеличению флуоресценции [13]. Стоит отметить, что однофлуорофорные индикаторы отличаются от двухфлуорофорных, которые изменяют конформацию белка от вытянутой до более компактной при связывании кальция, что приводит к увеличению эффективности FRET от ФБ с короткой длиной волны к ФБ с большей длиной волны [21]. У однофлуорофорных индикаторов использовался дизайн, отличный от подхода, основанного на FRET, в случае Camgaroo кальций-связывающий домен CaM вставлен в середину YFP. В дальнейшем, индикаторы Pericams [76] и G-CaMP (позднее сокращенный до GCaMP) [77] расширили эту концепцию, присоединив CaM к С-концу и CaM-связывающий пептид к N-концу кругового пермутированного флуоресцентного белка [75, 78]. На текущий момент, основными представителями однофлуорофорных ГеККИ являются индикаторы семейства GCaMP [77], которые все чаще используются для визуализации кальция in vivo [39, 79, 80].
Как отмечалось выше, дизайн GCaMP основан на молекулярном слиянии быстрой кальций-связывающей части белка (CaM), кальмодулин-связывающего пептида из киназы легкой цепи миозина гладких мышц (RS20, также известного как M13 [77]) и одного кругового пермутированного флуоресцентного белка, как правило, зеленого (cpEGFP) [44]. При связывании с Ca2+ мотив CaM (может связывать до четырех ионов кальция) вызывает конформационные изменения, которые обеспечивают связывание с M13. Эти взаимодействия вызывают депротонирование хромофора, что приводит к изменению спектральных свойств и увеличению интенсивности излучения сигнала (рис. 1) [41, 81, 82].
Рис. 1.
Схематическое изображение однофлуорофорных генетически кодированных индикаторов кальция. Связывание Ca2+ (красные кружки) с кальций-связывающим доменом (CaM–M13) увеличивает флуоресценцию кругового пермутированного флуоресцентного белка (изображен зеленым оттенком).
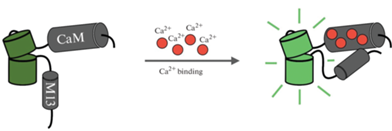
Оптический сигнал, создаваемый GCaMP, пропорционален доле молекул индикатора, связанных с Ca2+ [44]. Флуоресценцию GCaMP обычно регистрируют при возбуждении на длине волны 470–480 нм при однофотонном, 920–940 нм при двухфотонном и ~1300 нм при трехфотонном возбуждении [47, 83–85]. При этом скорость флуоресцентных конформаций GCaMP может быть ограничена не только скоростью связывания кальция, но и скоростью взаимодействия CaM и M13 [44]. Также стоит отметить, что для Ca2+-индикаторов время включения обычно быстрее, чем затухания, поскольку первое обычно ограничено связыванием Ca2+ с индикатором, а второе – буферизацией и откреплением Ca2+ от индикатора [63].
ОСНОВНЫЕ ПАРАМЕТРЫ, ХАРАКТЕРИЗИРУЮЩИЕ ОДНОФЛУОРОФОРНЫЕ ГЕККИ
Важными характеристиками ГеККИ являются: динамический диапазон, соотношение сигнал–шум, отношение сигнала к базовому уровню, константа диссоциации и коэффициент Хилла.
Динамический диапазон (D) является одним из наиболее важных характеристик ГеККИ, который для однофлуорофорных индикаторов определяется по формуле:
где D – динамический диапазон, а Fmax и Fmin– соответственно максимальная и минимальная интенсивности флуоресценции, полученные в условиях Ca2+-насыщения или Ca2+-деплеции.Динамический диапазон также может быть выражен в процентах и может быть определен следующим образом:
Как видно из уравнения, двукратное увеличение интенсивности флуоресценции соответствует 100%-ному увеличению динамического диапазона.
Отношение сигнала к базовому уровню (SBR) или мера сигнала, сообщаемого индикатором во время изменения внутриклеточной концентрации кальция для интенсиометрического индикатора, определяется отношением флуоресцентного ответа к базовой флуоресценции [19]:
Как правило, способность индикатора регистрировать Ca2+-транзиент на фоне флуоресценции определяется на основе соотношения сигнал–шум (SNR), характерного для индикатора, которое вычисляется как отношение между сигналом флуоресценции (ΔF) и дробным шумом базовой флуоресценции [19]:
где $S{{D}_{{{{F}_{0}}}}}$ – стандартное отклонение базовой флуоресценции (до изменения концентрации кальция в клетке).Еще одной важной характеристикой интенсиометрических индикаторов является фракционное насыщение ($\theta $) [86], которое может быть определено следующим образом:
Реакции, способствующие изменению оптических свойств ГеККИ при связывании Ca2+ в условиях равновесия, могут быть описаны с помощью константы диссоциации (Kd):
где koff и kon– скорости ассоциации и диссоциации между лигандом и рецептором, эти параметры определяют, насколько быстро и эффективно индикатор связывается и высвобождает ионы кальция в ответ на изменения концентрации кальция. Значение Kd позволяет качественно сравнивать ГеККИ между собой и дает представление о приблизительном диапазоне концентраций Ca2+, в котором могут функционировать ГеККИ. Чем ниже значение Kd, тем меньше концентрация кальция, необходимая для насыщения индикатора, и, следовательно, тем более чувствительным является индикатор к изменениям концентрации кальция в клетке.Связь между фракционным насыщением индикатора, концентрацией кальция и Kd индикатора выражается с помощью уравнения Хилла [87]: $\theta = ~\,\,\frac{{{{{\left[ {{\text{C}}{{{\text{a}}}^{{{\text{2 + }}}}}} \right]}}^{n}}}}{{\left( {{{{\left[ {{\text{C}}{{{\text{a}}}^{{{\text{2 + }}}}}} \right]}}^{n}} + ~{{K}_{d}}} \right)~}},$
где n – коэффициент Хилла, а остальные переменные имеют то же значение, что описано выше. Константа Хилла показывает кривизну зависимости между концентрацией кальция и сигналом (флуоресценцией индикатора). Коэффициент Хилла может принимать значения больше, меньше или равные 1. Если коэффициент Хилла равен 1, то зависимость между концентрацией кальция и сигналом линейна. Если коэффициент Хилла больше 1, то зависимость имеет более крутой наклон и является сигналом с усилением, что может быть полезно для измерения кальция в более высоких концентрациях. Если коэффициент Хилла меньше 1, то зависимость имеет более плавный наклон и является сигналом с ослаблением, что может быть полезно для измерения кальция в более низких концентрациях.
Для определения коэффициента Хилла и константы диссоциации проводят калибровку ГеККИ. Калибровка представляет собой процесс определения связи между флуоресцентным сигналом, который генерирует кальциевый индикатор, и концентрацией ионов кальция. В процессе калибровки ГеККИ проводятся эксперименты, в которых измеряют флуоресцентный сигнал индикатора при различных известных концентрациях кальция. На основе полученных данных строится калибровочная кривая, которая позволяет обратно преобразовать флуоресцентный сигнал ГеККИ в соответствующую концентрацию ионов кальция, необходимую для дальнейшего измерения кальциевой активности в биологических системах.
Для калибровки используют очищенный белок ГеККИ [12]. Титрование кальция проводят по нескольким точкам, изменяя концентрацию кальция от 100 нМ до 10 мМ. Для этого разбавляют очищенный белок ГеККИ в соотношении 1 : 50 в буферном растворе, содержащем 30 мМ MOPS (pH 7.2) и 100 мМ KCl. В зависимости от требуемой концентрации свободного кальция, в буфер добавляют либо 10 мМ EGTA (без свободного кальция), либо 10 мМ CaEGTA (~39 мкМ свободного кальция), смешивают в различных пропорциях. Далее, используя спектрофотометр с микропланшетным ридером, например, Safire 2 (Tecan), регистрируют флуоресценцию индикатора при определенных длинах волн. Например, для GCaMP возбуждение осуществляется на длине волны 485 нм, а эмиссия измеряется на длине волны 510 нм. Затем полученные данные используются для построения калибровочных кривых связывания индикатора с кальцием. Для аппроксимации этих кривых используют сигмоидальные функции, после чего определяют коэффициент Хилла и Kd.
Наряду с калибровкой ГеККИ определяют значения ряда параметров, таких как константа диссоциации по кислотности (pKa), коэффициент экстинкции (ε) и квантовый выход (Φ).
Определение константы диссоциации по кислотности (pKa) осуществляется путем титрования по pH. pKa представляет собой значение pH, при котором половина молекул ГеККИ находится в протонированной форме (несвязанной с кальцием) и половина в депротонированной форме (связанной с кальцием). При pH ниже pKa большая часть ГеККИ будет находиться в протонированной форме, а при pH выше pKa – в депротонированной форме. Для определения константы диссоциации очищенный белок ГеККИ разбавляют в pH-буферах, содержащих либо 5 мМ CaCl2 для индикатора в связанном с кальцием состоянии, либо 5 мМ EGTA для ГеККИ в несвязанном с кальцием состоянии соответственно. Буферы готовят таким образом, чтобы получить двенадцать различных значений pH в диапазоне от 4.5 до 10.5 [12]. Значения pKa определяют по точке перегиба кривой флуоресценции в зависимости от pH. Определение pKa для ГеККИ позволяет подобрать оптимальное pH, при котором будет достигнута наилучшая флуоресцентная активность и стабильность индикатора.
Коэффициент экстинкции (ε) ГеККИ определяется как отношение поглощенной интенсивности света к концентрации индикатора в растворе и позволяет оценить, насколько эффективно флуорофор поглощает свет определенной длины волны. Коэффициент экстинкции ГеККИ важен для вычисления концентрации кальция в клетке на основе изменений интенсивности флуоресценции, вызванных взаимодействием индикатора с кальцием. Чем больше значение коэффициента экстинкции, тем больше света поглощается флуорофором, и тем сильнее может быть его флуоресцентный сигнал. Значение коэффициента экстинкции можно определить экспериментально, используя известную концентрацию ГеККИ и значение поглощенной интенсивности света, измеренное с помощью спектрофотометра. Для определения коэффициента экстинкции индикатора в связанном и несвязанном с кальцием состояниях, очищенный белок ГеККИ растворяют в 50 мМ MOPS, 100 мМ KCl (pH 7.2), содержащем либо 5 мМ CaCl2, либо 10 мМ EGTA соответственно. Затем измеряется поглощенная интенсивность света при соответствующей длине волны, например, для GCaMP – при 497 или 505 нм [12]. Концентрацию ГеККИ в растворе определяют путем денатурации белка в 0.1 М NaOH с использованием коэффициента экстинкции 44 000 М−1 см−1 при 447 нм для денатурированного хромофора GFP [12].
Квантовый выход (Φ) определяется как отношение количества испускаемых фотонов (флуоресцентного сигнала) к количеству поглощенных фотонов (возбуждающего света) и показывает, насколько эффективно ГеККИ преобразует энергию поглощенного света в флуоресцентный сигнал. Квантовый выход измеряется в процентах или в долях единицы (от 0 до 1). Например, если квантовый выход составляет 0.6, это означает, что ГеККИ испускает примерно 60% фотонов в виде флуоресцентного сигнала, а остаток энергии теряется в виде нагрева или других процессов. Чем выше значение квантового выхода, тем более эффективно флуорофор преобразует поглощенные фотоны в испускаемую флуоресценцию. Для определения квантового выхода, очищенный белок ГеККИ, насыщенный кальцием, растворяют в 50 мМ MOPS, 100 мМ KCl и 5 мМ CaCl2. Затем, с помощью спектрофотометра анализируется количество испускаемых и поглощенных фотонов. Для GCaMP пики возбуждения/эмиссии без кальция и с кальцием составляют 505 нм/517 нм и 497 нм/515 нм соответственно [12].
РАЗВИТИЕ GCaMP
Оригинальная версия сенсора GCaMP [77] была итеративно усовершенствована несколькими группами исследователей путем многочисленных раундов мутагенеза и селекции (рис. 2, табл. 1) [13]. В результате появились следующие версии сенсора: GCaMP1.6 [88], GCamp2 [89], GCamp3 [41], высокочувствительные GCaMP, такие как GCaMP-HS [83], Fast-GCaMPs [90], GCaMP4.1 [91], GCamp5 [92], GCamp6 (GCaMP6s, GCaMP6m и GCaMP6f) [12], jGCamp7 (т.е. jGCaMP7s, jGCaMP7f, j-GCaMP7b и jGCaMP7c) [93], jGCaMP8 (jGCaMP8f, jGCaMP8m и jGCaMP8s) [94]. Самыми востребованными версиями GCaMP за исключением последних модификаций индикатора были GCaMP3, GCaMP5, GCaMP6 [95].
Рис. 2.
Хронология развития основных представителей семейства GCaMP по датам соответствующих публикаций.
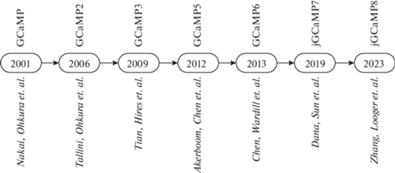
Таблица 1.
Характеристики основных представителей семейства GCaMP
| Вариант GCaMP | Кальций-связывающий домен | Kd (нМ) | Коэффициент Хилла | Источник |
|---|---|---|---|---|
| GCaMP | CaM-M13 | 235 | 3.3 | [77] |
| G-CaMP2 | 146 | 3.8 | [89] | |
| GCaMP3 | 330 | 3.2 | [100] | |
| GCaMP5A | 307 | 2.7 | [92] | |
| GCaMP5G | 460 | 2.5 | ||
| GCaMP5K | 189 | 3.8 | ||
| GCaMP6s | 144 | 2.9 | [12] | |
| GCaMP6m | 167 | 2.96 | ||
| GCaMP6f | 375 | 2.27 | ||
| jGCaMP7f | 174 | 2.3 | [93] | |
| jGCaMP7s | 68 | 2.49 | ||
| jGCaMP7c | 298 | 2.44 | ||
| jGCaMP7b | 82 | 3.06 | ||
| jGCaMP8f | CaM-eNOS | 334 | 2.08 | [94] |
| jGCaMP8m | 108 | 1.92 | ||
| jGCaMP8s | 46 | 2.2 |
Следует отметить, что некоторые научные группы используют отдельную систему нумерации для вариантов GCaMP [95], которые включают GCaMP6 [96], G-CaMP7 [96], GCaMP7a [97], G-CaMP7.09 [98] и GCaMP8 [96], GCaMP9a [99], GCamp-X [65]. Однако эти GCaMP являются постепенным усовершенствованием не серии GCaMP6, а более ранних версий [12].
GCaMPs продемонстрировали свое применение для обнаружения ПД и определенных субклеточных событий in vivo. Одно из их первых применений in vivo – визуализация одорантных ответов у мух, где использовался GCaMP1 [39]. С тех пор GCaMPs использовались для визуализации активности в мозге червей и рыб, что позволило выявить специфические нейроны, участвующие в принятии решений или обучении [101–104]. Регистрация ПД в одной и той же популяции нейронов с течением времени с помощью GCaMP позволила выявить процесс, при котором определенные нейроны в мозге начинают работать вместе для выполнения определенной функции (рекрутирование) во время обучения у мыши [105–107]. GCaMP также использовались для локализации активности в определенных постсинаптических и пресинаптических компартментах [108].
С момента появления GCaMP2 ГеККИ на его основе стали регулярно использоваться для визуализации in vivo, поскольку GCaMP2, в отличие от предыдущих версий GCaMP, обладал пониженной чувствительностью к рН и был полностью функционален при 37°C [109]. Благодаря этим особенностям GCaMP2 использовался для отслеживания пресинаптических кальциевых транзиентов, отдельных спайков и больших популяций нейронов [12, 110]. В результате GCaMP2 послужил шаблоном для создания высокочувствительных GCaMP с улучшенным временным и пространственным разрешением для in vivo исследований [73, 83].
Значительный прогресс был достигнут в версии GCaMP3, которая обладала повышенной яркостью, динамическим диапазоном и сродством к Ca2+, что позволило регистрировать повышение Ca2+, вызванное отдельными потенциалами действия in vitro [41]. GCaMP3 был первым вариантом ГеККИ, для которого были созданы трансгенные мыши, обеспечивающие неинвазивную, хроническую экспрессию c использованием Cre-рекомбиназы [111, 112]. Наличие таких мышей значительно расширило возможности использования GCaMP3. Кроме того, GCaMP3 был использован для обнаружения активности в больших популяциях нейронов в моторной коре [41], бочкообразной коре [113] и гиппокампе [80] мышей. Долгосрочная визуализация с использованием GCaMP3 выявила изменения в нейронной сети, связанные с обучением, in vivo [114]. GCaMP3 также использовался для исследования активности популяции нейронов в слое 5 (L5) соматосенсорной коры взрослой мыши [115], изучения сетчатки мыши на клеточном и субклеточном уровнях [116], зрительного тракта зебрафиш [117] и зрительно-моторного пути дрозофилы [118] и др. Тем не менее, на тот момент времени, GCaMP3 оставался несовершенным ГеККИ, т.к. не позволял стабильно регистрировать одиночные ПД in vivo и отставал по скорости обнаружения активных клеток от показателей синтетических кальциевых сенсоров [41].
На основе GCaMP3 был создан набор вариантов GCaMP5 путем целевого мутагенеза линкерных последовательностей, соединяющих M13/CaM с cpGFP [81, 92, 119]. Двенадцать GCaMP5 (5A, 5D, 5G, 5K, 5L и др.) были систематически охарактеризованы в культивируемых нейронах, астроцитах, а также в Caenorhabditis elegans, дрозофиле, зебрафиш и мыши в различных условиях. Все варианты GCaMP5 продемонстрировали улучшенный динамический диапазон по сравнению с GCaMP3, среди которых отдельно отмечались варианты GCaMP5A, GCaMP5G и GCaMP5K [95]. В результате улучшения характеристик, GCaMP5 использовался для локализации одиночных событий высвобождения нейромедиаторов в нервно-мышечном соединении мухи, показав, что спонтанные и вызванные события происходят на разных бутонах [120].
Впоследствии на основе GCaMP5 был создан GCaMP6 путем модификаций аминокислот между cpGFP-CaM и M13-CaM [12]. На диссоциированной культуре нейронов были проверены 447 вариантов GCaMP, в результате чего были получены три сверхчувствительных варианта GCaMP6 с различной кинетикой: GCaMP6s, GCaMP6m и GCaMP6f, характеризующиеся медленной, средней и быстрой кинетикой соответственно. По сравнению с GCaMP5, эти сенсоры имели 1.1–1.6-кратное увеличение динамического диапазона и в три раза более высокую аффинность связывания Ca2+. Кроме того, GCaMP6f на то время демонстрировал самую быструю кинетику среди всего семейства GCaMP благодаря мутации в домене M13-CaM. Поколение генетически кодированных сенсоров GCaMP6 (GCaMP6s, GCaMP6m и GCaMP6f) стало самым востребованным и произвело прорыв в области прижизненной кальциевой визуализации [95]. Так, например, GCaMP6 использовался для регистрации кальциевых транзиентов от дендритных отростков возбуждающих тормозных интернейронов, расположенных в первичной зрительной коре [12]. В экспериментах с использованием GCaMP6s было показано, что разные отростки на одном и том же нейроне зрительной коры отвечают на зрительные стимулы разной ориентации [12]. С помощью GCaMP6s было обнаружено, что локальные дендритные шипики коррелируют с синаптическим потенцированием в нейронах коры головного мозга [121] и приобретением специфичности места в нейронах гиппокампа [122]. Кроме того, GCaMP6s использовали для обнаружения ориентационной избирательности отдельных аксональных бутонов в проекциях зрительной коры [123]. В свою очередь, GCaMP6f использовался для изучения нейронных сетей у бодрствующих свободно передвигающихся мышей с высоким временным разрешением путем однофотонной широкопольной визуализации с помощью миниатюрных микроскопов [124]. В целом, можно сказать, что варианты GCaMP6 активно использовались для визуализации активности нейронов у бодрствующих животных [125–130].
Дальнейшая оптимизация GCaMP6 привела к созданию серий jGCaMP7 (Janelia GCaMP7) [93] и jGCaMP8 (Janelia GCaMP8) [94]. jGCaMP7 был создан путем модификаций аминокислот между cpGFP-CaM, M13-CaM и M13-cpGFP. На диссоциированной культуре нейронов были проверены 662 варианта GCaMP [93]. В результате были отобраны следующие варианты GCaMP7 с различными свойствами: jGCaMP7s (чувствительный), jGCaMP7b (яркий), jGCaMP7c (контрастный), jGCaMP7f (быстрый). jGCaMP7s показал более низкую константу диссоциации (более высокое сродство) для связывания Ca2+ по сравнению с GCaMP6s (68 против 147 нМ). jGCaMP7f, jGCaMP7b и jCaMP7s продемонстрировали более высокую базовую флуоресценцию, а также большую амплитуду ΔF/F0 по сравнению с GCaMP6f. Кинетика jGCaMP7f оказалась быстрее, чем у других сенсоров jGCaMP7, и была сравнима с GCaMP6f. jGCaMP7s, в отличие от других версий jGCaMP7, имел более медленное время затухания. jGCaMP7b демонстрировал 50%-ное увеличение базового уровня флуоресценции по сравнению с GCaMP6s. В сравнении с семейством GCaMP6, индикаторы jGCaMP7 обеспечивали улучшенное обнаружение отдельных спайков (jGCaMP7s,f), визуализацию в нейритах и нейропиле (jGCaMP7b) и позволяли отслеживать большие популяции нейронов с помощью двухфотонной (jGCaMP7s,f) или широкопольной (jGCaMP7c) визуализации [93].
jGCaMP8 был разработан путем замены пептида M13 на пептид эндотелиальной синтазы оксида азота (eNOS) [94]. Такая конструкция придавала сенсору более быструю кинетику, что улучшало отслеживание потенциалов действия. На диссоциированной культуре нейронов были проверены 32 варианта GCaMP. В результате было отобрано три перспективных варианта: jGCaMP8s (быстрое нарастание, медленное затухание, высокая чувствительность), jGCaMP8f (быстрое нарастание и быстрое затухание) и jGCaMP8m (быстрое нарастание и среднее затухание). Все индикаторы jGCaMP8 имели более быстрое время нарастания (~10 мс) и спада (~50–200 мс) флуоресценции, чем время нарастания (~50–200 мс) и спада (~150–500 мс) у GCaMP6 [12]. Варианты jGCaMP8 позволяли регистрировать отдельные спайки в нейронах с частотой до 50 Гц. ΔF/F0 и SNR для всех индикаторов jGCaMP8 были больше, чем для jGCaMP7, при этом jGCaMP8f в три быстрее jGCaMP7f. Семейство jGCaMP8 идеально подходит для отслеживания активности быстроспайкующих интернейронов. Из-за быстрого времени затухания флуоресценции индикаторы jGCaMP8 более предпочтительны для визуализации с более высокой частотой дискретизации, чем широко используемые индикаторы jGCaMP7s и GCaMP6s.
ЦВЕТОВЫЕ РАЗНОВИДНОСТИ GCAMP
С появлением GCaMP6, jGCaMP7 и jGCaMP8 оптимизация ГеККИ типа GCaMP, по крайней мере для зеленой флуоресцентной разновидности, казалось, достигла стадии, когда дальнейшее улучшение труднодостижимо или невозможно. Тем не менее были созданы варианты интенсиометрических кальциевых сенсоров с ФБ отличными от GFP [68, 69, 131]. В результате путем замены cpEGFP на cpmApple и cpmRuby были созданы красные флуоресцентные Ca2+-индикаторы, такие как R-GECO [68] и RCaMP [119]. Впоследствии в семействе GECO были созданы многоцветные варианты: B-GECO (синие), Y-GECO (желтые) и NIR-GECO (инфракрасные) [132–135]. Зеленые флуоресцентные Ca2+-индикаторы, как правило, работают лучше всего, но наличие других цветовых вариантов позволяет пользователям одновременно получать изображения Ca2+-активности различных типов клеток в разных цветах или выбирать ГеККИ, спектрально совместимые с другими индикаторами, используемыми в эксперименте. Стоит отметить, что флуоресцентные индикаторы со смещенными в красную область спектрами возбуждения обладают сниженной фоновой автофлуоресценцией, меньшей фототоксичностью и лучшим соотношением сигнал–шум благодаря меньшему поглощению красных длин волн в тканях мозга [68].
Сенсоры семейства GECO [68] были созданы на основе GCaMP3 путем случайного мутагенеза. GECO позволяли маркировать несколько клеточных популяций разными цветами и осуществлять визуализацию в глубоких слоях мозга. Однако эти варианты страдали слабой интенсивностью и ограниченным контрастом для визуализации in vivo.
Аналогично GCaMP, R-GECO был впоследствии оптимизирован и сконструирован в R-CaMP1.07, R-CaMP2, XCaMP-R, jRGECO и K-GECO1 [69, 131, 136]. Так, например, R-CaMP2 был создан путем замены фрагмента M13 в R-CaMP1.07 (в то время обладавшего наивысшими характеристиками красного ГеККИ) на пептид ckkap из киназы CaMK (CaMKK) из 150 мутаций, которые были внесены в домен cpRFP-CaM. R-CaMP2 имел в три раза более высокую чувствительность к Ca2+ (Kd = 70 нМ) и коэффициент Хилла 1.2. Этот подход был также использован при разработке красного флуоресцентного Ca2+-индикатора K-GECO1, который включал ckkap в качестве CaM-связывающего пептида и демонстрировал улучшенную чувствительность, быструю кинетику и линейный ответ на спайки [136].
Отдельно стоит упомянуть более поздний набор многоцветных ГеККИ, созданный на основе GCaMP4.1 и получивший название X-CaMP (синий: XCaMP-B, зеленый: XCaMP-G, желтый: XCaMP-Y, красный: XCaMP-R) [131]. В серии XCaMP был использован подход, аналогичный созданию R-CaMP2. В отличие от классических представителей GCaMP, в XCaMP пептид M13 был заменен на пептид ckkap из CaMKK. Предыдущие биохимические анализы показали, что CaMKK обладает высоким сродством к Ca2+ и низким коэффициентом Хилла (∼1), т.е. линейно реагирует на кальций [131]. XCaMPs позволяет обнаруживать одиночные спайки в течение 3–10 мс после начала спайка, что особенно важно для оценки быстроспайкующих интернейронов [131], в то время как у многих представителей семейства GCaMP коэффициенты Хилла для связывания Ca2+ больше единицы, что делает их менее чувствительными к одному или нескольким спайкам [22]. Кроме того, повышенная базовая флуоресценция X-CaMP облегчает использование кальциевой визуализации для оценки подпороговых событий в шипиках и синаптических терминалях [131]. Новые цветовые варианты (синий, желтый и красный) в серии X-CaMP облегчают многоцветную визуализацию с 1-фотонным или 2-фотонным возбуждением флуоресценции.
На сегодняшний день многоцветная визуализация с использованием кальциевых сенсоров становится все более актуальна. Например, использование различных многоцветных сенсоров может способствовать раскрытию взаимодействия между нейронами и астроцитами в процессе формирования нейронных связей [95]. Так, использование двухцветной одновременной двухфотонной визуализации нейронов и астроцитов в зрительной коре бодрствующей мыши имело решающее значение для обнаружения зрительного ответа астроцитов, в частности того, как этот ответ связан с активностью соседних нейронов [137].
Еще одним преимуществом многоцветной Ca2+-визуализации является одновременная визуализация пре- и постсинаптических, субклеточных компартментов нейронов из перекрывающихся аксонов и дендритов или отростков астроцитов [69, 131, 138]. Двухцветные одновременные наблюдения в слое 1 бочковой коры мыши с использованием XCaMP-R в дендритах пирамидальных нейронов и XCaMP-Y в аксоне SST-нейронов позволили охарактеризовать обработку информации через синапсы [131]. Постсинаптические Ca2+-события были подавлены только в дендритных сегментах с перекрывающимися аксонами. Дендритные события также достоверно подавлялись за секунду до и после активации аксона. В совокупности эти результаты определяют пространственные и временные масштабы локального дендритного торможения в апикальных отростках пирамидальных нейронов.
Кроме всего выше сказанного, отдельно стоит упомянуть использование многоцветных ГеККИ в оптогенетических экспериментах. Оптогенетическая модуляция в сочетании с мониторингом клеточной активности на основе GCaMP позволяет использовать полностью оптическую электрофизиологию (ОЭ). Первоначальные усилия по созданию ОЭ были затруднены из-за спектрального разделения индикаторов. Например, зеленые GCaMP не могут быть использованы в клетках, экспрессирующих каналородопсин-2 (ChR2), галородопсин (HR) или архродопсин (Arch), поскольку спектр возбуждающего света ГеККИ перекрывается со спектрами активации светочувствительных каналов [44]. Примером красного ГеККИ, который удалось использовать в сочетании с оптогенетикой, является вариант jRGECO1a [93]. Попытки еще больше сдвинуть спектр возбуждения ГЕККИ в красную область привели к созданию индикатора кальция ближнего инфракрасного (NIR) диапазона, NIR-GECO1, который состоит из кальцийсвязывающих доменов CaM и M13, вставленных в основанный на бактериофитохроме NIR-флуоресцентный белок mIFP [134]. Дополнительные спектры возбуждения ГеККИ обеспечивают гибкость в инструментарии оптофизиологии, позволяя анализировать различные нейронные ансамбли [68].
Расширение цветового спектра значительно увеличило потенциал ГеККИ в многоцветной визуализации. Применение красных флуоресцентных индикаторов позволили уменьшить рассеяние ткани, фототоксичность и фоновую флуоресценцию, способствуя визуализации в глубоких слоях мозга. Применение синих и желтых вариантов ГеККИ, помимо красных и зеленых, позволили исследователям проводить одновременную визуализацию различных нейронных ансамблей, а также комбинировать применение генетически кодируемых кальциевых индикаторов с оптогенетическими инструментами [119].
ЗАКЛЮЧЕНИЕ
Применение генетически кодируемых кальциевых индикаторов представляет собой важный инструмент для изучения нейрональных процессов и нейронных ансамблей в реальном времени. В этом обзоре была представлена необходимая информация об однофлуорофорных ГеККИ: их преимуществах и недостатках, разновидностях, особенностях текущего использования и перспективах применения для визуализации нейронной активности. Один из главных плюсов ГеККИ заключается в том, что они позволяют получать детальную информацию о кальциевых сигналах в реальном времени. Современные тенденции совершенствования ГеККИ направлены на расширение их спектральных свойств и способности регистрировать высокочастотную спайковую активность путем модификаций домена CaM-M13. Благодаря многоцветной in vivo визуализации с использованием ГеККИ и ее комбинированию с такими методами, как оптогенетика, нейробиологи вносят дополнительный вклад в наше понимание функций мозга. Очевидно, что в ближайшие годы мы увидим новые версии ГеККИ, учитывая большое количество CaM-связывающих белков (более 300) [139], которые могут быть полезны для дальнейшего усовершенствования этих индикаторов.
Список литературы
Grienberger C, Konnerth A (2012) Imaging calcium in neurons. Neuron 73: 862–885. https://doi.org/10.1016/j.neuron.2012.02.011
Russell JT (2011) Imaging calcium signals in vivo: a powerful tool in physiology and pharmacology. Br J Pharmacol 163: 1605–1625. https://doi.org/10.1111/j.1476-5381.2010.00988.x
Ghosh A, Greenberg ME (1995) Calcium signaling in neurons: molecular mechanisms and cellular consequences. Science 268: 239–247. https://doi.org/10.1126/science.7716515
Kawamoto EM, Vivar C, Camandola S (2012) Physiology and pathology of calcium signaling in the brain. Front Pharmacol 3: 61. https://doi.org/10.3389/fphar.2012.00061
Tsien RY, Pozzan T, Rink TJ (1982) Calcium homeostasis in intact lymphocytes: cytoplasmic free calcium monitored with a new, intracellularly trapped fluorescent indicator. J Cell Biol 94: 325–334. https://doi.org/10.1083/jcb.94.2.325
Grynkiewicz G, Poenie M, Tsien RY (1985) A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem 260: 3440–3450.
Tsien RY (1981) A non-disruptive technique for loading calcium buffers and indicators into cells. Nature 290: 527–528. https://doi.org/10.1038/290527a0
Inoue M (2021) Genetically encoded calcium indicators to probe complex brain circuit dynamics in vivo. Neurosci Res 169: 2–8. https://doi.org/10.1016/j.neures.2020.05.013
Reck-Peterson SL, Derr ND, Stuurman N (2010) Imaging single molecules using total internal reflection fluorescence microscopy (TIRFM). Cold Spring Harb Protoc 2010: pdb top73. https://doi.org/10.1101/pdb.top73
Toseland CP (2013) Fluorescent labeling and modification of proteins. J Chem Biol 6: 85–95. https://doi.org/10.1007/s12154-013-0094-5
Renz M (2013) Fluorescence microscopy-a historical and technical perspective. Cytometry A 83: 767–779. https://doi.org/10.1002/cyto.a.22295
Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger L, Svoboda K, Kim DS (2013) Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499: 295–300. https://doi.org/10.1038/nature12354
Lin MZ, Schnitzer MJ (2016) Genetically encoded indicators of neuronal activity. Nat Neurosci 19: 1142–1153. https://doi.org/10.1038/nn.4359
Carafoli E (2003) The calcium-signalling saga: tap water and protein crystals. Nat Rev Mol Cell Biol 4: 326–332. https://doi.org/10.1038/nrm1073
Pchitskaya E, Popugaeva E, Bezprozvanny I (2018) Calcium signaling and molecular mechanisms underlying neurodegenerative diseases. Cell Calcium 70: 87–94. https://doi.org/10.1016/j.ceca.2017.06.008
Supnet C, Bezprozvanny I (2010) Neuronal calcium signaling, mitochondrial dysfunction, and Alzheimer’s disease. J Alzheimers Dis 20 Suppl 2: S487–S498. https://doi.org/10.3233/JAD-2010-100306
Sabatini BL, Svoboda K (2000) Analysis of calcium channels in single spines using optical fluctuation analysis. Nature 408: 589–593. https://doi.org/10.1038/35046076
Kim TH, Schnitzer MJ (2022) Fluorescence imaging of large-scale neural ensemble dynamics. Cell 185: 9–41. https://doi.org/10.1016/j.cell.2021.12.007
Yasuda R, Nimchinsky EA, Scheuss V, Pologruto TA, Oertner TG, Sabatini BL, Svoboda K (2004) Imaging calcium concentration dynamics in small neuronal compartments. Sci STKE 2004: pl5. https://doi.org/10.1126/stke.2192004pl5
Tank DW, Sugimori M, Connor JA, Llinas RR (1988) Spatially resolved calcium dynamics of mammalian Purkinje cells in cerebellar slice. Science 242: 773–777. https://doi.org/10.1126/science.2847315
Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY (1997) Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388: 882–887. https://doi.org/10.1038/42264
Rose T, Goltstein PM, Portugues R, Griesbeck O (2014) Putting a finishing touch on GECIs. Front Mol Neurosci 7: 88. https://doi.org/10.3389/fnmol.2014.00088
Ashley CC (1969) Aequorin-monitored calcium transients in single Maia muscle fibres. J Physiol 203: 32P–33P.
Ashley CC, Ridgway EB (1968) Simultaneous recording of membrane potential, calcium transient and tension in single muscle fibers. Nature 219: 1168–1169. https://doi.org/10.1038/2191168a0
Shimomura O, Johnson FH, Saiga Y (1962) Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol 59: 223–239. https://doi.org/10.1002/jcp.1030590302
Mank M, Griesbeck O (2008) Genetically encoded calcium indicators. Chem Rev 108: 1550–1564. https://doi.org/10.1021/cr078213v
Heim R, Tsien RY (1996) Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol 6: 178–182. https://doi.org/10.1016/s0960-9822(02)00450-5
Porumb T, Yau P, Harvey TS, Ikura M (1994) A calmodulin-target peptide hybrid molecule with unique calcium-binding properties. Protein Eng 7: 109–115. https://doi.org/10.1093/protein/7.1.109
Romoser VA, Hinkle PM, Persechini A (1997) Detection in living cells of Ca2+-dependent changes in the fluorescence emission of an indicator composed of two green fluorescent protein variants linked by a calmodulin-binding sequence. A new class of fluorescent indicators. J Biol Chem 272: 13270–13274. https://doi.org/10.1074/jbc.272.20.13270
Siegel MS, Isacoff EY (1997) A genetically encoded optical probe of membrane voltage. Neuron 19: 735–741. https://doi.org/10.1016/s0896-6273(00)80955-1
Miesenbock G, De Angelis DA, Rothman JE (1998) Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394: 192–195. https://doi.org/10.1038/28190
Thestrup T, Litzlbauer J, Bartholomaus I, Mues M, Russo L, Dana H, Kovalchuk Y, Liang Y, Kalamakis G, Laukat Y, Becker S, Witte G, Geiger A, Allen T, Rome LC, Chen TW, Kim DS, Garaschuk O, Griesinger C, Griesbeck O (2014) Optimized ratiometric calcium sensors for functional in vivo imaging of neurons and T lymphocytes. Nat Methods 11: 175–182. https://doi.org/10.1038/nmeth.2773
Palmer AE, Tsien RY (2006) Measuring calcium signaling using genetically targetable fluorescent indicators. Nat Protoc 1: 1057–1065. https://doi.org/10.1038/nprot.2006.172
Mank M, Santos AF, Direnberger S, Mrsic-Flogel TD, Hofer SB, Stein V, Hendel T, Reiff DF, Levelt C, Borst A, Bonhoeffer T, Hubener M, Griesbeck O (2008) A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nat Methods 5: 805–811. https://doi.org/10.1038/nmeth.1243
Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A (2004) Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci U S A 101: 10554–10559. https://doi.org/10.1073/pnas.0400417101
Kerr R, Lev-Ram V, Baird G, Vincent P, Tsien RY, Schafer WR (2000) Optical imaging of calcium transients in neurons and pharyngeal muscle of C. elegans. Neuron 26: 583–594. https://doi.org/10.1016/s0896-6273(00)81196-4
Boulin T, Hobert O (2012) From genes to function: the C. elegans genetic toolbox. Wiley Interdiscip Rev Dev Biol 1: 114–137. https://doi.org/10.1002/wdev.1
Higashijima S, Masino MA, Mandel G, Fetcho JR (2003) Imaging neuronal activity during zebrafish behavior with a genetically encoded calcium indicator. J Neurophysiol 90: 3986–3997. https://doi.org/10.1152/jn.00576.2003
Wang JW, Wong AM, Flores J, Vosshall LB, Axel R (2003) Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell 112: 271–282. https://doi.org/10.1016/s0092-8674(03)00004-7
Hasan MT, Friedrich RW, Euler T, Larkum ME, Giese G, Both M, Duebel J, Waters J, Bujard H, Griesbeck O, Tsien RY, Nagai T, Miyawaki A, Denk W (2004) Functional fluorescent Ca2+ indicator proteins in transgenic mice under TET control. PLoS Biol 2: e163. https://doi.org/10.1371/journal.pbio.0020163
Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, Bargmann CI, Jayaraman V, Svoboda K, Looger LL (2009) Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods 6: 875–881. https://doi.org/10.1038/nmeth.1398
Heider B, Nathanson JL, Isacoff EY, Callaway EM, Siegel RM (2010) Two-photon imaging of calcium in virally transfected striate cortical neurons of behaving monkey. PLoS One 5: e13829. https://doi.org/10.1371/journal.pone.0013829
Yin L, Masella B, Dalkara D, Zhang J, Flannery JG, Schaffer DV, Williams DR, Merigan WH (2014) Imaging light responses of foveal ganglion cells in the living macaque eye. J Neurosci 34: 6596–665. https://doi.org/10.1523/JNEUROSCI.4438-13.2014
Mollinedo-Gajate I, Song C, Knopfel T (2019) Genetically Encoded Fluorescent Calcium and Voltage Indicators. Handb Exp Pharmacol 260: 209–229. https://doi.org/10.1007/164_2019_299
Mao T, O’Connor DH, Scheuss V, Nakai J, Svoboda K (2008) Characterization and subcellular targeting of GCaMP-type genetically-encoded calcium indicators. PLoS One 3: e1796. https://doi.org/10.1371/journal.pone.0001796
Stosiek C, Garaschuk O, Holthoff K, Konnerth A 2003) In vivo two-photon calcium imaging of neuronal networks. Proc Natl Acad Sci U S A 100: 7319–7324. https://doi.org/10.1073/pnas.1232232100
Cui G, Jun SB, Jin X, Pham MD, Vogel SS, Lovinger DM, Costa RM (2013) Concurrent activation of striatal direct and indirect pathways during action initiation. Nature 494: 238–242. https://doi.org/10.1038/nature11846
Bozza T, McGann JP, Mombaerts P, Wachowiak M (2004) In vivo imaging of neuronal activity by targeted expression of a genetically encoded probe in the mouse. Neuron 42: 9–21. https://doi.org/10.1016/s0896-6273(04)00144-8
Kaifosh P, Lovett-Barron M, Turi GF, Reardon TR, Losonczy A (2013) Septo-hippocampal GABAergic signaling across multiple modalities in awake mice. Nat Neurosci 16: 1182–1184. https://doi.org/10.1038/nn.3482
Knopfel T (2012) Genetically encoded optical indicators for the analysis of neuronal circuits. Nat Rev Neurosci 13: 687–700. https://doi.org/10.1038/nrn3293
Steinmetz NA, Aydin C, Lebedeva A, Okun M, Pachitariu M, Bauza M, Beau M, Bhagat J, Bohm C, Broux M, Chen S, Colonell J, Gardner R J, Karsh B, Kloosterman F, Kostadinov D, Mora-Lopez C, O’Callaghan J, Park J, Putzeys J, Sauerbrei B, van Daal R JJ, Vollan AZ, Wang S, Welkenhuysen M, Ye Z, Dudman JT, Dutta B, Hantman AW, Harris KD, Lee A, Moser EI, O’Keefe J, Renart A, Svoboda K, Hausser M, Haesler S, Carandini M, Harris T D (2021) Neuropixels 2.0: A miniaturized high-density probe for stable, long-term brain recordings. Science 372. https://doi.org/10.1126/science.abf4588
Erofeev A, Antifeev I, Bolshakova A, Bezprozvanny I, Vlasova O (2022) In Vivo Penetrating Microelectrodes for Brain Electrophysiology. Sensors (Basel) 22. https://doi.org/10.3390/s22239085
Margolis DJ, Lutcke H, Schulz K, Haiss F, Weber B, Kugler S, Hasan MT, Helmchen F (2012) Reorganization of cortical population activity imaged throughout long-term sensory deprivation. Nat Neurosci 15: 1539–1546. https://doi.org/10.1038/nn.3240
Patriarchi T, Cho JR, Merten K, Howe MW, Marley A, Xiong WH, Folk RW, Broussard G J, Liang R, Jang MJ, Zhong H, Dombeck D, von Zastrow M, Nimmerjahn A, Gradinaru V, Williams JT, Tian L (2018) Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360. https://doi.org/10.1126/science.aat4422
Sabatini BL, Tian L (2020) Imaging Neurotransmitter and Neuromodulator Dynamics In Vivo with Genetically Encoded Indicators. Neuron 108: 17–32. https://doi.org/10.1016/j.neuron.2020.09.036
Emiliani V, Cohen AE, Deisseroth K, Hausser M (2015) All-Optical Interrogation of Neural Circuits. J Neurosci 35: 13917–13926. https://doi.org/10.1523/JNEUROSCI.2916-15.2015
Kuhn B, Ozden I, Lampi Y, Hasan MT, Wang SS (2012) An amplified promoter system for targeted expression of calcium indicator proteins in the cerebellar cortex. Front Neural Circuits 6: 49. https://doi.org/10.3389/fncir.2012.00049
Resendez SL, Jennings JH, Ung RL, Namboodiri VM, Zhou ZC, Otis JM, McHenry JA, Kosyk O, Stuber GD (2016) Visualization of cortical, subcortical and deep brain neural circuit dynamics during naturalistic mammalian behavior with head-mounted microscopes and chronically implanted lenses. Nat Protoc 11: 566–597. https://doi.org/10.1038/nprot.2016.021
Bedbrook CN, Deverman BE, Gradinaru V (2018) Viral Strategies for Targeting the Central and Peripheral Nervous Systems. Annu Rev Neurosci 41: 323–348. https://doi.org/10.1146/annurev-neuro-080317-062048
Haery L, Deverman BE, Matho KS, Cetin A, Woodard K, Cepko C, Guerin KI, Rego MA, Ersing I, Bachle SM, Kamens J, Fan M (2019) Adeno-Associated Virus Technologies and Methods for Targeted Neuronal Manipulation. Front Neuroanat 13: 93. https://doi.org/10.3389/fnana.2019.00093
Chan KY, Jang MJ, Yoo BB, Greenbaum A, Ravi N, Wu WL, Sanchez-Guardado L, Lois C, Mazmanian SK, Deverman BE, Gradinaru C (2017) Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat Neurosci 20: 1172–1179. https://doi.org/10.1038/nn.4593
Allen WE, Kauvar IV, Chen MZ, Richman EB, Yang SJ, Chan K, Gradinaru V, Deverman BE, Luo L, Deisseroth K (2017) Global Representations of Goal-Directed Behavior in Distinct Cell Types of Mouse Neocortex. Neuron 94: 891–907 e896. https://doi.org/10.1016/j.neuron.2017.04.017
Helmchen F, Imoto K, Sakmann B (1996) Ca2+ buffering and action potential-evoked Ca2+ signaling in dendrites of pyramidal neurons. Biophys J 70: 1069–1081. https://doi.org/10.1016/S0006-3495(96)79653-4
Helmchen F, Borst JG, Sakmann B (1997) Calcium dynamics associated with a single action potential in a CNS presynaptic terminal. Biophys J 72: 1458–1471. https://doi.org/10.1016/S0006-3495(97)78792-7
Yang Y, Liu N, He Y, Liu Y, Ge L, Zou L, Song S, Xiong W, Liu X (2018) Improved calcium sensor GCaMP-X overcomes the calcium channel perturbations induced by the calmodulin in GCaMP. Nat Commun 9: 1504. https://doi.org/10.1038/s41467-018-03719-6
Jurado LA, Chockalingam PS, Jrrett HW (1999) Apocalmodulin. Physiol Rev 79: 661–682. https://doi.org/10.1152/physrev.1999.79.3.661
Marvin JS, Borghuis BG, Tian L, Cichon J, Harnett MT, Akerboom J, Gordus A, Renninger SL, Chen TW, Bargmann CI, Orger MB, Schreiter ER, Demb JB, Gan WB, Hires SA, Looger LL (2013) An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat Methods 10: 162–170. https://doi.org/10.1038/nmeth.2333
Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, Nakano M, Abdelfattah AS, Fujiwara M, Ishihara T, Nagai T, Campbell RE (2011) An expanded palette of genetically encoded Ca2+ indicators. Science 333: 1888–1891. https://doi.org/10.1126/science.1208592
Dana H, Mohar B, Sun Y, Narayan S, Gordus A, Hasseman JP, Tsegaye G, Holt GT, Hu A, Walpita D, Patel R, Macklin JJ, Bargmann CI, Ahrens MB, Schreiter ER, Jayaraman V, Looger LL, Svoboda K, Kim DS (2016) Sensitive red protein calcium indicators for imaging neural activity. Elife 5. https://doi.org/10.7554/eLife.12727
Wu SY, Shen Y, Shkolnikov I, Campbell RE (2022) Fluorescent Indicators For Biological Imaging of Monatomic Ions. Front Cell Dev Biol 10: 885440. https://doi.org/10.3389/fcell.2022.885440
Shen Y, Wu SY, Rancic V, Aggarwal A, Qian Y, Miyashita SI, Ballanyi K, Campbell RE, Dong M (2019) Genetically encoded fluorescent indicators for imaging intracellular potassium ion concentration. Commun Biol 2: 18. https://doi.org/10.1038/s42003-018-0269-2
Helassa N, Podor B, Fine A, Torok (2016) Design and mechanistic insight into ultrafast calcium indicators for monitoring intracellular calcium dynamics. Sci Rep 6: 38276. https://doi.org/10.1038/srep38276
Badura A, Sun XR, Giovannucci A, Lynch LA, Wang SS (2014) Fast calcium sensor proteins for monitoring neural activity. Neurophotonics 1: 025008. https://doi.org/10.1117/1.NPh.1.2.025008
Bovetti S, Moretti C, Fellin T (2014) Mapping brain circuit function in vivo using two-photon fluorescence microscopy. Microsc Res Tech 77: 492–501. https://doi.org/10.1002/jemt.22342
Baird GS, Zacharias DA, Tsien RY (1999) Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci U S A 96: 11241–11246. https://doi.org/10.1073/pnas.96.20.11241
Nagai T, Sawano A, Park ES, Miyawaki A (2001) Circularly permuted green fluorescent proteins engineered to sense Ca2+. Proc Natl Acad Sci U S A 98: 3197–3202. https://doi.org/10.1073/pnas.051636098
Nakai J, Ohkura M, Imoto K (2001) A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nat Biotechnol 19: 137–141. https://doi.org/10.1038/84397
Souslova EA, Belousov VV, Lock JG, Stromblad S, Kasparov S, Bolshakov AP, Pinelis V G, Labas YA, Lukyanov S, Mayr LM, Chudakov DM (2007) Single fluorescent protein-based Ca2+ sensors with increased dynamic range. BMC Biotechnol 7: 37. https://doi.org/10.1186/1472-6750-7-37
Fletcher ML, Masurkar AV, Xing J, Imamura F, Xiong W, Nagayama S, Mutoh H, Greer C, Knopfel T, Chen WR (2009) Optical imaging of postsynaptic odor representation in the glomerular layer of the mouse olfactory bulb. J Neurophysiol 102: 817–830. https://doi.org/10.1152/jn.00020.2009
Dombeck DA, Harvey CD, Tian L, Looger LL, Tank DW (2010) Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat Neurosci 13: 1433–1440. https://doi.org/10.1038/nn.2648
Akerboom J, Rivera JD, Guilbe MM, Malave EC, Hernandez HH, Tian L, Hires SA, Marvin JS, Looger LL, Schreiter ER (2009) Crystal structures of the GCaMP calcium sensor reveal the mechanism of luorescence signal change and aid rational design. J Biol Chem 284: 6455–6464. https://doi.org/10.1074/jbc.M807657200
Iseppon F, Linley JE, Wood JN (2022) Calcium imaging for analgesic drug discovery. Neurobiol Pain 11: 100083. https://doi.org/10.1016/j.ynpai.2021.100083
Muto A, Ohkura M, Kotani T, Higashijima S, Nakai J, Kawakami K (2011) Genetic visualization with an improved GCaMP calcium indicator reveals spatiotemporal activation of the spinal motor neurons in zebrafish. Proc Natl Acad Sci U S A 108: 5425 –5430. https://doi.org/10.1073/pnas.1000887108
Chen Q, Cichon J, Wang W, Qiu L, Lee SJ, Campbell NR, Destefino N, Goard MJ, Fu Z, Yasuda R, Looger LL, Arenkiel BR, Gan WB, Feng G (2012) Imaging neural activity using Thy1-GCaMP transgenic mice. Neuron 76: 297–308. https://doi.org/10.1016/j.neuron.2012.07.011
Ouzounov DG, Wang T, Wu C, Xu C (2019) GCaMP6 DeltaF/F dependence on the excitation wavelength in 3-photon and 2-photon microscopy of mouse brain activity. Biomed Opt Express 10: 3343–3352. https://doi.org/10.1364/BOE.10.003343
Hires SA, Tian L, Looger LL (2008) Reporting neural activity with genetically encoded calcium indicators. Brain Cell Biol 36: 69–86. https://doi.org/10.1007/s11068-008-9029-4
Perez Koldenkova V, Nagai T (2013) Genetically encoded Ca(2+) indicators: properties and evaluation. Biochim Biophys Acta 1833: 1787–1797. https://doi.org/10.1016/j.bbamcr.2013.01.011
Ohkura M, Matsuzaki M, Kasai H, Imoto K, Nakai J (2005) Genetically encoded bright Ca2+ probe applicable for dynamic Ca2+ imaging of dendritic spines. Anal Chem 77: 5861–5869. https://doi.org/10.1021/ac0506837
Tallini YN, Ohkura M, Choi BR, Ji G, Imoto K, Doran R, Lee J, Plan P, Wilson J, Xin HB, Sanbe A, Gulick J, Mathai J, Robbins J, Salama G, Nakai J, Kotlikoff MI (2006) Imaging cellular signals in the heart in vivo: Cardiac expression of the high-signal Ca2+ indicator GCaMP2. Proc Natl Acad Sci U S A 103: 4753–4758. https://doi.org/10.1073/pnas.0509378103
Sun XR, Badura A, Pacheco DA, Lynch LA, Schneider ER, Taylor MP, Hogue IB, Enquist LW, Murthy M, Wang SS (2013) Fast GCaMPs for improved tracking of neuronal activity. Nat Commun 4: 2170. https://doi.org/10.1038/ncomms3170
Shindo A, Hara Y, Yamamoto TS, Ohkura M, Nakai J, Ueno N (2010) Tissue-tissue interaction-triggered calcium elevation is required for cell polarization during Xenopus gastrulation. PLoS One 5: e8897. https://doi.org/10.1371/journal.pone.0008897
Akerboom J, Chen TW, Wardill TJ, Tian L, Marvin JS, Mutlu S, Calderon NC, Esposti F, Borghuis BG, Sun XR, Gordus A, Orger MB, Portugues R, Engert F, Macklin JJ, Filosa A, Aggarwal A, Kerr RA, Takagi R, Kracun S, Shigetomi E, Khakh BS, Baier H, Lagnado L, Wang SS, Bargmann CI, Kimmel BE, Jayaraman V, Svoboda K, Kim DS, Schreiter ER, Looger LL (2012) Optimization of a GCaMP calcium indicator for neural activity imaging. J Neurosci 32: 13819–13840. https://doi.org/10.1523/JNEUROSCI.2601-12.2012
Dana H, Sun Y, Mohar B, Hulse BK, Kerlin AM, Hasseman JP, Tsegaye G, Tsang A, Wong A, Patel R, Macklin JJ, Chen Y, Konnerth A, Jayaraman V, Looger LL, Schreiter ER, Svoboda K, Kim DS (2019) High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat Methods 16: 649–657. https://doi.org/10.1038/s41592-019-0435-6
Zhang Y, Looger LL (2023) Fast and sensitive GCaMP calcium indicators for neuronal imaging. J Physiol 10: 1113/JP283832. https://doi.org/10.1113/JP283832
Broussard GJ, Liang R, Tian L (2014) Monitoring activity in neural circuits with genetically encoded indicators. Front Mol Neurosci 7: 97. https://doi.org/10.3389/fnmol.2014.00097
Ohkura M, Sasaki T, Sadakari J, Gengyo-Ando K, Kagawa-Nagamura Y, Kobayashi C, Ikegaya Y, Nakai J (2012) Genetically encoded green fluorescent Ca2+ indicators with improved detectability for neuronal Ca2+ signals. PLoS One 7: e51286. https://doi.org/10.1371/journal.pone.0051286
Muto A, Ohkura M, Abe G, Nakai J, Kawakami K (2013) Real-time visualization of neuronal activity during perception. Curr Biol 23: 307–311. https://doi.org/10.1016/j.cub.2012.12.040
Shiba Y, Gomibuchi T, Seto T, Wada Y, Ichimura H, Tanaka Y, Ogasawara T, Okada K, Shiba N, Sakamoto K, Ido D, Shiina T, Ohkura M, Nakai J, Uno N, Kazuki Y, Oshimura M, Minami I, Ikeda U (2016) Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature 538: 388–391. https://doi.org/10.1038/nature19815
Sakamoto M, Inoue M, Takeuchi A, Kobari S, Yokoyama T, Horigane SI, Takemoto-Kimura S, Abe M, Sakimura K, Kano M, Kitamura K, Fujii H, Bito H (2022) A Flp-dependent G-CaMP9a transgenic mouse for neuronal imaging in vivo. Cell Rep Methods 2: 100168. https://doi.org/10.1016/j.crmeth.2022.100168
Helassa N, Zhang XH, Conte I, Scaringi J, Esposito E, Bradley J, Carter T, Ogden D, Morad M, Torok K (2015) Fast-Response Calmodulin-Based Fluorescent Indicators Reveal Rapid Intracellular Calcium Dynamics. Sci Rep 5: 15978. https://doi.org/10.1038/srep15978
Ahrens MB, Li JM, Orger MB, Robson DN, Schier AF, Engert F, Portugues R (2012) Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature 485: 471–477. https://doi.org/10.1038/nature11057
Dunn TW, Mu Y, Narayan S, Randlett O, Naumann EA, Yang CT, Schier AF, Freeman J, Engert F, Ahrens M (2016) Brain-wide mapping of neural activity controlling zebrafish exploratory locomotion. Elife 5: e12741. https://doi.org/10.7554/eLife.12741
Venkatachalam V, Ji N, Wang X, Clark C, Mitchell JK, Klein M, Tabone CJ, Florman J, Ji H, Greenwood J, Chisholm AD, Srinivasan J, Alkema M, Zhen M, Samuel AD (2016) Pan-neuronal imaging in roaming Caenorhabditis elegans. Proc Natl Acad Sci U S A 113: E1082–E1088. https://doi.org/10.1073/pnas.1507109113
Nguyen JP, Shipley FB, Linder AN, Plummer GS, Liu M, Setru SU, Shaevitz JW, Leifer AM (2016) Whole-brain calcium imaging with cellular resolution in freely behaving Caenorhabditis elegans. Proc Natl Acad Sci U S A 113: E1074–E1081. https://doi.org/10.1073/pnas.1507110112
Peters AJ, Chen SX, Komiyama T (2014) Emergence of reproducible spatiotemporal activity during motor learning. Nature 510: 263–267. https://doi.org/10.1038/nature13235
Ziv Y, Burns L, Cocker ED, Hamel EO, Ghosh KK, Kitch LJ, El Gamal A, Schnitzer MJ (2013) Long-term dynamics of CA1 hippocampal place codes. Nat Neurosci 16: 264–266. https://doi.org/10.1038/nn.3329
Lovett-Barron M, Kaifosh P, Kheirbek MA, Danielson N, Zaremba JD, Reardon TR, Turi GF, Hen R, Zemelman BV, Losonczy A (2014) Dendritic inhibition in the hippocampus supports fear learning. Science 343: 857–863. https://doi.org/10.1126/science.1247485
Siegel F, Lohmann C (2013) Probing synaptic function in dendrites with calcium imaging. Exp Neurol 242: 27–32. https://doi.org/10.1016/j.expneurol.2012.02.007
Kotlikoff MI (2007) Genetically encoded Ca2+ indicators: using genetics and molecular design to understand complex physiology. J Physiol 578: 55–67. https://doi.org/10.1113/jphysiol.2006.120212
Diez-Garcia J, Matsushita S, Mutoh H, Nakai J, Ohkura M, Yokoyama J, Dimitrov D, Knopfel T (2005) Activation of cerebellar parallel fibers monitored in transgenic mice expressing a fluorescent Ca2+ indicator protein. Eur J Neurosci 22: 627–635. https://doi.org/10.1111/j.1460-9568.2005.04250.x
Zariwala HA, Borghuis BG, Hoogland TM, Madisen L, Tian L, De Zeeuw CI, Zeng H, Looger LL, Svoboda K, Chen TW (2012) A Cre-dependent GCaMP3 reporter mouse for neuronal imaging in vivo. J Neurosci 32: 3131–3141. https://doi.org/10.1523/JNEUROSCI.4469-11.2012
Paukert M, Agarwal A, Cha J, Doze VA, Kang JU, Bergles DE (2014) Norepinephrine controls astroglial responsiveness to local circuit activity. Neuron 82: 1263–1270. https://doi.org/10.1016/j.neuron.2014.04.038
O'Connor DH, Peron SP, Huber D, Svoboda K (2010) Neural activity in barrel cortex underlying vibrissa-based object localization in mice. Neuron 67: 1048–1061. https://doi.org/10.1016/j.neuron.2010.08.026
Huber D, Gutnisky DA, Peron S, O’Connor DH, Wiegert JS, Tian L, Oertner TG, Looger LL, Svoboda K (2012) Multiple dynamic representations in the motor cortex during sensorimotor learning. Nature 484: 473–478. https://doi.org/10.1038/nature11039
Mittmann W, Wallace DJ, Czubayko U, Herb JT, Schaefer AT, Looger LL, Denk W, Kerr JN (2011) Two-photon calcium imaging of evoked activity from L5 somatosensory neurons in vivo. Nat Neurosci 14: 1089–1093. https://doi.org/10.1038/nn.2879
Borghuis BG, Tian L, Xu Y, Nikonov SS, Vardi N, Zemelman BV, Looger LL (2011) Imaging light responses of targeted neuron populations in the rodent retina. J Neurosci 31: 2855–2867. https://doi.org/10.1523/JNEUROSCI.6064-10.2011
Del Bene F, Wyart C, Robles E, Tran A, Looger L, Scott EK, Isacoff EY, Baier H (2010) Filtering of visual information in the tectum by an identified neural circuit. Science 330: 669–673. https://doi.org/10.1126/science.1192949
Chiappe ME, Seelig JD, Reiser MB, Jayaraman V (2010) Walking modulates speed sensitivity in Drosophila motion vision. Curr Biol 20: 1470–1475. https://doi.org/10.1016/j.cub.2010.06.072
Akerboom J, Carreras Calderon N, Tian L, Wabnig S, Prigge M, Tolo J, Gordus A, Orger MB, Severi KE, Macklin JJ, Patel R, Pulver SR, Wardill TJ, Fischer E, Schuler C, Chen T W, Sarkisyan KS, Marvin JS, Bargmann CI, Kim DS, Kugler S, Lagnado L, Hegemann P, Gottschalk A, Schreiter ER, Looger LL (2013) Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front Mol Neurosci 6: 2. https://doi.org/10.3389/fnmol.2013.00002
Melom JE, Akbergenova Y, Gavornik JP, Littleton JT (2013) Spontaneous and evoked release are independently regulated at individual active zones. J Neurosci 33: 17253–17263. https://doi.org/10.1523/JNEUROSCI.3334-13.2013
Cichon J, Gan WB (2015) Branch-specific dendritic Ca(2+) spikes cause persistent synaptic plasticity. Nature 520: 180–185. https://doi.org/10.1038/nature14251
Sheffield ME, Dombeck DA (2015) Calcium transient prevalence across the dendritic arbour predicts place field properties. Nature 517: 200–204. https://doi.org/10.1038/nature13871
Sun W, Tan Z, Mensh BD, Ji N (2016) Thalamus provides layer 4 of primary visual cortex with orientation- and direction-tuned inputs. Nat Neurosci 19: 308–315. https://doi.org/10.1038/nn.4196
Aharoni D, Khakh BS, Silva AJ, Golshani P (2019) All the light that we can see: a new era in miniaturized microscopy. Nat Methods 16: 11–13. https://doi.org/10.1038/s41592-018-0266-x
Boyd AM, Kato HK, Komiyama T, Isaacson JS (2015) Broadcasting of cortical activity to the olfactory bulb. Cell Rep 10: 1032–1039. https://doi.org/10.1016/j.celrep.2015.01.047
Dana H, Chen TW, Hu A, Shields BC, Guo C, Looger LL, Kim DS, Svoboda K (2014) Thy1-GCaMP6 transgenic mice for neuronal population imaging in vivo. PLoS One 9: e108697. https://doi.org/10.1371/journal.pone.0108697
Hinckley CA, Alaynick WA, Gallarda BW, Hayashi M, Hilde KL, Driscoll SP, Dekker JD, Tucker HO, Sharpee TO, Pfaff S (2015) Spinal Locomotor Circuits Develop Using Hierarchical Rules Based on Motorneuron Position and Identity. Neuron 87: 1008–1021. https://doi.org/10.1016/j.neuron.2015.08.005
Theis L, Berens P, Froudarakis E, Reimer J, Roman Roson M, Baden T, Euler T, Tolias A S, Bethge ER (2016) Benchmarking Spike Rate Inference in Population Calcium Imaging. Neuron 90: 471–482. https://doi.org/10.1016/j.neuron.2016.04.014
Heckscher ES, Zarin AA, Faumont S, Clark MQ, Manning L, Fushiki A, Schneider-Mizell CM, Fetter RD, Truman JW, Zwart M F, Landgraf M, Cardona A, Lockery SR, Doe CQ (2015) Even-Skipped(+) Interneurons Are Core Components of a Sensorimotor Circuit that Maintains Left-Right Symmetric Muscle Contraction Amplitude. Neuron 88: 314–329. https://doi.org/10.1016/j.neuron.2015.09.009
Grover D, Katsuki T, Greenspan RJ (2016) Flyception: imaging brain activity in freely walking fruit flies. Nat Methods 13: 569–572. https://doi.org/10.1038/nmeth.3866
Inoue M, Takeuchi A, Manita S, Horigane SI, Sakamoto M, Kawakami R, Yamaguchi K, Otomo K, Yokoyama H, Kim R, Yokoyama T, Takemoto-Kimura S, Abe M, Okamura M, Kondo Y, Quirin S, Ramakrishnan C, Imamura T, Sakimura K, Nemoto T, Kano M, Fujii H, Deisseroth K, Kitamura K, Bito H (2019) Rational Engineering of XCaMPs, a Multicolor GECI Suite for In Vivo Imaging of Complex Brain Circuit Dynamics. Cell 177: 1346–1360 e1324. https://doi.org/10.1016/j.cell.2019.04.007
Mohr MA, Bushey D, Aggarwal A, Marvin JS, Kim JJ, Marquez EJ, Liang Y, Patel R, Macklin JJ, Lee CY, Tsang A, Tsegaye G, Ahrens AM, Chen JL, Kim DS, Wong AM, Looger LL, Schreiter ER, Podgorski K (2020) jYCaMP: an optimized calcium indicator for two-photon imaging at fiber laser wavelengths. Nat Methods 17: 694–697. https://doi.org/10.1038/s41592-020-0835-7
Shcherbakova DM (2021) Near-infrared and far-red genetically encoded indicators of neuronal activity. J Neurosci Methods 362: 109314. https://doi.org/10.1016/j.jneumeth.2021.109314
Qian Y, Piatkevich KD, Mc Larney B, Abdelfattah AS, Mehta S, Murdock MH, Gottschalk S, Molina RS, Zhang W, Chen Y, Wu J, Drobizhev M, Hughes TE, Zhang J, Schreiter ER, Shoham S, Razansky D, Boyden ES, Campbell RE (2019) A genetically encoded near-infrared fluorescent calcium ion indicator. Nat Methods 16: 171–174. https://doi.org/10.1038/s41592-018-0294-6
Shemetov AA, Monakhov MV, Zhang Q, Canton-Josh JE, Kumar M, Chen M, Matlashov ME, Li X, Yang W, Nie L, Shcherbakova DM, Kozorovitskiy Y, Yao J, Ji N, Verkhusha VV (2021) A near-infrared genetically encoded calcium indicator for in vivo imaging. Nat Biotechnol 39: 368–377. https://doi.org/10.1038/s41587-020-0710-1
Shen Y, Dana H, Abdelfattah AS, Patel R, Shea J, Molina R S, Rawal B, Rancic V, Chang YF, Wu L, Chen Y, Qian Y, Wiens MD, Hambleton N, Ballanyi K, Hughes TE, Drobizhev M, Kim DS, Koyama M, Schreiter ER, Campbell R E (2018) A genetically encoded Ca(2+) indicator based on circularly permutated sea anemone red fluorescent protein eqFP578. BMC Biol 16: 9. https://doi.org/10.1186/s12915-018-0480-0
Sonoda K, Matsui T, Bito H, Ohki K (2018) Astrocytes in the mouse visual cortex reliably respond to visual stimulation. Biochem Biophys Res Commun 505: 1216–1222. https://doi.org/10.1016/j.bbrc.2018.10.027
Bindocci E, Savtchouk I, Liaudet N, Becker D, Carriero G, Volterra A (2017) Three-dimensional Ca2+ imaging advances understanding of astrocyte biology. Science 356. https://doi.org/10.1126/science.aai8185
Yap KL, Kim J, Truong K, Sherman M, Yuan T, Ikura M (2000) Calmodulin target database. J Struct Funct Genomics 1: 8–14. https://doi.org/10.1023/a:1011320027914
Дополнительные материалы отсутствуют.
Инструменты
Российский физиологический журнал им. И.М. Сеченова


