Биологические мембраны: Журнал мембранной и клеточной биологии, 2022, T. 39, № 6, стр. 419-430
Рецепторы SARS-CoV-2 и их участие в инфицировании клеток
П. П. Авдонин a, Е. Ю. Рыбакова a, С. К. Труфанов a, П. В. Авдонин a, *
a Институт биологии развития им. Н.К. Кольцова РАН
119334 Москва, Россия
* E-mail: pvavdonin@yandex.ru
Поступила в редакцию 10.07.2022
После доработки 30.07.2022
Принята к публикации 01.08.2022
- EDN: TQESHR
- DOI: 10.31857/S0233475522060032
Аннотация
Пандемия новой коронавирусной инфекции (COVID-19), вызванная вирусом SARS-CoV-2, многократно превзошла по масштабам эпидемии, вызванные SARS-CoV и MERS-CoV. Причиной этого стало наличие в белковой последовательности SARS-CoV-2 участков, обеспечивающих взаимодействие с более широким спектром рецепторных белков на поверхности клеток хозяина. В обзоре рассмотрены как уже известные рецепторы, общие для SARS-CoV и SARS-CoV-2, так и новые рецепторы, специфичные для SARS-CoV-2.
ВВЕДЕНИЕ
C начала XXI века три коронавируса преодолели межвидовой барьер и стали причиной смертельной пневмонии у людей: коронавирус тяжелого острого респираторного синдрома (SARS-CoV), ближневосточный коронавирус респираторного синдрома (MERS-CoV) и коронавирус тяжелого острого респираторного синдрома 2 (SARS-CoV-2) [1]. Первые два из этих вирусов вызвали локальные эпидемии, тогда как появление SARS-CoV-2 привело к пандемии, которая продолжается уже около трех лет. По данным Всемирной организации здравоохранения (https://covid19.who.int), к началу августа 2022 г. в мире подтверждено более 570 млн случаев коронавирусной инфекции. Скончалось около 6.4 млн человек. Процент смертности от COVID-19 после начала массовой вакцинации существенно снизился, но наличие постковидных осложнений, так называемого постковидного синдрома, остается серьезной проблемой. Появление все новых вариантов SARS-CoV-2 по-прежнему вызывает большую озабоченность и внимание всего мира. Накопление мутаций в геноме вируса обусловлено непрерывной репликацией вируса в организме человека на фоне несвоевременной элиминации вирионов, связанной в основном с запоздалым клиническим лечением и отсутствием специфических и эффективных препаратов. Эти мутации могут привести к изменениям эпитопов и к снижению аффинности соответствующих антител, генерируемых вакцинами. Поэтому важно разработать универсальные, специфические и эффективные препараты против имеющихся и возможных будущих вариантов коронавируса.
Способом защиты от коронавирусной инфекции может быть подавление транспорта вирусных частиц в клетки. На первом этапе инфицирования происходит связывание S-белка шипиков короны с рецепторами на мембране клеток-мишеней, расщепление S-белка на фрагменты S1 и S2, после чего происходит слияние вирусной мембраны с плазматической мембраной клетки либо эндоцитоз вирусных частиц [2–4]. После этого мРНК SARS-CoV-2 заходит внутрь клетки. Каноническим рецептором SARS-CoV-2 является ангиотензинпревращающий фермент 2 (ACE2), работающий в паре с мембранной протеазой TRMPSS2 [2]. Механизм проникновения в клетки с помощью ACE2 и TRMPSS2 был открыт ранее для вируса SARS-CoV. При анализе патогенеза новой коронавирусной инфекции выявилась парадоксальная ситуация – поражение вирусом SARS-CoV-2 клеток и органов во многих случаях не коррелировало с экспрессией в них ACE2 [5]. Это стимулировало поиск рецепторных и вспомогательных белков, специфичных именно для SARS-CoV-2. В результате исследований последних трех лет подтверждено участие в инфицировании клеток SARS-CoV-2 белковых мишеней SARS-CoV, но также открыты новые рецепторы, которые обуславливают более высокую распространенность COVID-19 (рис. 1). В настоящем обзоре кратко охарактеризованы мишени коронавируса SARS-CoV-2 на клеточной поверхности и мембранные протеазы, активирующие S-белок. Кратко рассмотрена их роль в инфицировании разных тканей и клеток организма.
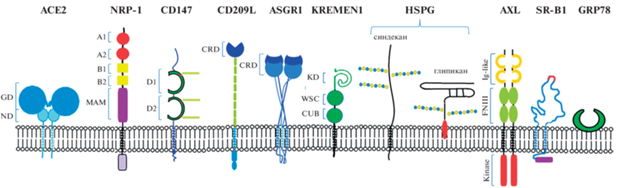
Рис. 1.
Рецепторы и корецепторы, задействованные в инфицировании клеток коронавирусом SARS-CoV2: ангиотензинпревращающий фермент 2 (ACE2), нейропилин-1 (NRP-1), CD147 или базигин, CD209L или L-SIGN, рецептор 1 асиалогликопротеинов (ASGR1), белок KREMEN1 (kringle containing transmembrane protein 1 – трансмембранный белок, содержащий крингл-домен), гепарансульфатные протеогликаны (HSPG) синдекан и глипикан, рецепторная тирозинкиназа AXL, скэвенджер-рецептор типа 1 липопротеинов высокой плотности (SR-B1), GRP78 (glucose-regulated protein 78 – регулируемый глюкозой белок 78). Показаны домены рецепторных белков – GD (globular domain – глобулярный домен), ND (neck domain – соединительный домен), MAM (meprin, A-5 protein, receptor protein-tyrosine phosphatase µ), иммуноглобулиновые домены (D1 и D2), CRD (carbohydrate recognition domain – узнающий углеводы домен), KD (kringle domain – крингл или крендель домен), WSC (cell wall integrity and stress response component – компонент целостности клеточной стенки и ответа на стресс), CUB (complement C1r/C1s, Uegf, Bmp1), Ig-like (иммуноглобулин-подобный домен), FNIII (fibronectin type III – домен фибронектина III типа), kinase (тирозинкиназа), гликозильные остатки на молекуле CD147 и гепарансульфаты синдеканов и глипиканов.
ПРАЙМИРОВАНИЕ СПАЙКОВОГО БЕЛКА КОРОНАВИРУСА И ПРОНИКНОВЕНИЕ SARS-COV-2 В КЛЕТКИ
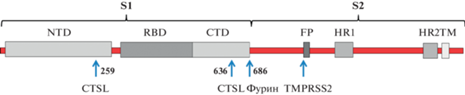
S-белок относится к вирусным fusion-белкам класса I, для которых характерна тримерная структура [6, 7]. Масса мономера составляет 180–200 кДа, к белку присоединено большое число полисахаридов [8]. Строение мономера S-белка показано на рис. 2. Пептидная цепь S-белка SARS-CoV-2 образована 1273 а.о. и включает сигнальный пептид (аминокислоты 1–13), расположенный на N-конце, и фрагменты S1 (14–685 а.о.) и S2 (686–1273 а.о.). В последовательности S1 имеется N-концевой домен (14–305 а.о.) и рецептор-связывающий домен (RBD – receptor binding domain, 319–541 а.о.), который непосредственно взаимодействует с ACE2. RBD может поворачиваться как на шарнире, переходя из компактной (положение “down”) в раскрытую (положение “up”) конформацию и обратно и при этом обнажая или скрывая мотив RBM (receptor binding motive), который связывается с рецепторным белком – ACE2 [9]. На поверхности вириона SARS-CoV-2 домен RBD S-белка спонтанно принимает обе эти конформации, и их соотношение может варьировать [10]. Далее после домена RBD расположен С-терминальный домен и участок, соединяющий S1 и S2. В состав фрагмента S2 входят fusion-пептид (FP) (788–806 а.о.), последовательность гептапептидных повторов 1 (HR1) (912–984 а.о.), HR2 (1163–1213 а.о.), трансмембранный домен TM и цитоплазматический домен (1237–1273 а.о.) [8].
Рис. 2.
Структура S-белка коронавируса SARS-CoV-2. NTD (N-terminal domain – N-концевой домен), RBD (receptor binding domain – рецептор-связывающий домен), CTD (C-terminal domain – С-концевой домен), FP (fusion peptide – пептид слияния), TM (transmembrane domain – трансмембранный домен), HR1 и HR2 (heptapeptide repeats 1 and 2 – гептапептидные повторы 1 и 2), CTPL (cathepsin L – катепсин L), TMPRSS2 (transmembrane protease, serine 2 – трансмембранная сериновая протеаза 2) Стрелками показаны точки протеолиза S-белка.
Для того, чтобы содержимое вирусной частицы SARS-CoV-2 поступило в цитоплазму клетки, необходима активация S-белка, так называемое праймирование. В результате протеолитического расщепления образуются субъединицы S1 и S2 [11]. Субъединица S1 отвечает за связывание с ACE2, S2 за слияние с мембраной клетки – мишени коронавируса. После праймирования домен RBD приобретает открытую конформацию (положение “up”), при которой он способен связываться рецептором [3]. Снимается также пространственное ограничительное влияние S1 на S2. Имеющиеся данные говорят о том, что процесс праймирования идет следующим образом (рис. 3): мембранная препротеинконвертаза фурин расщепляет белок S на S1- и S2-фрагменты справа от участка, содержащего основные аминокислоты R-X-(K/R)-R↓ (сайт протеолиза Q677TNSPRRAR-SV687, обозначаемый S1/S2). Фурин экпрессируется повсеместно в клетках организма. Наличие сайта протеолитического расщепления фурином считается основным фактором вирулентности высокопатогенных штаммов таких вирусов, как ВИЧ, респираторно-синцитиальный вирус, вирус кори. Этот сайт есть у SARS-CoV-2, но отсутствует у SARS-CoV [12]. Он появился в результате инсерции фрагмента PRRA в полипептидную цепь коронавируса [11]. Далее после действия на S-белок фурина происходит расщепление фрагмента S2 протеазой TMPRSS2 по сайту S2' (DPSKPSKR↓SFIED) на N-конце S2-субъединицы [11, 13]. TMPRSS2 расщепляет S-белки как SARS-CoV, так и SARS-CoV-2. Показана колокализация TMPRSS2 с ACE2 [14]. Интересно, что TMPRSS2 расщепляет также пептидную цепь ACE2 и тем самым приводит к отделению ACE2 от мембраны.
Рис. 3.
Праймирование S-белка коронавируса SARS-CoV-2 и слияние вирусной и клеточной мембран. НR1 и HR2 – гептапептидные повторы 1 и 2.
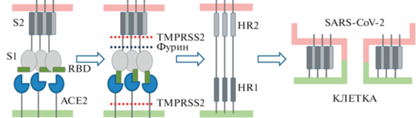
В результате протеолиза по сайту S2' открывается N-концевой гидрофобный пептид FP (fusion peptide) субъединицы S2. FP прикрепляется к мембране инфицируемой клетки, далее происходит “слипание” альфа-спиральных участков HR1 и HR2, субъединица S2 сокращается и таким образом подтягивает вирусную частицу к поверхности клетки, плазматическая мембрана и мембрана вируса сливаются, образуется контакт между содержимым вирусной частицы и цитоплазмой, и происходит поступление вирусной РНК внутрь клетки (рис. 3). Благодаря тримерной структуре в “подтягивании” вируса к клетке участвуют одновременно три S2-субъединицы. При этом образуется пучок из шести альфа-спиралей. В присоединении вируса к клетке задействованы, по всей вероятности, не одна, а несколько молекул S-белка разных шипиков короны.
Расщепление S-белка коронавируса SARS-CoV-2 на S1- и S2-субъединицы может осуществлять протеаза катепсин L [15]. Zhao и соавт. [16] выявили консервативные сайты протеолиза S-белка катепсином L в положениях 259 и 636 (рис. 2), имеющиеся у всех вариантов SARS-CoV-2 (включая вариант Omicron), и показали, что протеолиз по этим точкам способствует принятию рецептор-связывающим доменом (RBD) активированной конформации (положение “up”), при которой он присоединяется к ACE2 и заражает клетки. В экспериментах по заражению клеток была использована псевдовирусная конструкция SARS-CoV-2. По данным [17], праймирование S-белка катепсином L происходит при эндоцитозе вирусных частиц, в отличие от активации S-белка протеазой TMPRSS2, когда идет слияние вирусной и плазматической мембран. Эндоцитоз коронавируса SARS-CoV-2 осуществляется по клатрин-зависимому механизму [4]. Крайне важно в плане возможного лечения COVID-19 учитывать, что одновременное воздействие ингибиторов сериновой и цистеиновой протеаз TMPRSS2 (камостат мезилата или нафамостат мезилата) и катепсина L (гидроксихлорахина), которые входят в список известных лекарств, может синергично подавлять заражение клеток коронавирусом SARS-CoV-2 за счет блокирования обоих путей транспорта вирионов в клетки [17]. С помощью группового анализа дифференциальной экспрессии (genomic-guided gene set enrichment analysis – GSEA) эстрадиол и ретиноевая кислота также были идентифицированы как возможные фармпрепараты, способные подавлять активность TMPRSS2 и катепсина L и тем самым снижать транспорт SARS-CoV-2 в клетки [18]. Аналогичным образом, сетевой анализ перепрофилирования лекарств выявил возможность применения циклоспорина А, эстрадиола и кальцитриола в качестве потенциальных лекарств, подавляющих оба пути инфицирования клеток коронавирусом [19].
Поступление частиц SARS-CoV-2 в клетки происходит двумя путями – в результате эндоцитоза или слияния мембраны вирусной оболочки с плазматической мембраной клетки с высвобождением мРНК вируса в цитоплазму [20]. На вопрос о роли каждого из этих механизмов транспорта вирионов пока нет ясного ответа. В отношении SARS-CoV есть данные, что в легких инфицирование клеток через слияние мембран на 2–3 порядка эффективнее эндоцитоза благодаря предварительной активации S-белка экстраклеточными протеазами трипсином и термолизином [21]. Транспортный путь SARS-CoV-2 в клетки зависит от относительной экспрессии протеаз на плазматической мембране [17].
АНГИОТЕНЗИНПРЕВРАЩАЮЩИЙ ФЕРМЕНТ 2 (ACE2)
Роль ACE2 в качестве рецептора для коронавирусного S-белка первоначально была показана для SARS-CoV [22, 23], а позднее также для SARS-CoV-2 [2, 24, 25]. Домен RBD S1-субъединицы связывается с N-концевым пептидазным доменом ACE2 [26]. ACE2 открыт в 2000 г., его транскрипты обнаружены в сердце, почках и семенниках, а гистохимический анализ показал экспрессию в эндотелии коронарных и почечных сосудов и в тубулярном эпителии почек [27, 28]. Согласно атласу белков человека, ACE2 в наибольших количествах экспрессируется в кишечнике, почках и семенниках и в более низких концентрациях в легких и сердце [29]. В сосудистой системе наиболее высокая экспрессия ACE2 была выявлена в эндотелии микрососудов легких, а самая низкая – в эндотелии лимфатических сосудов [30]. Хотя инфекция SARS-CoV-2 в основном проявляется симптомами поражения респираторной системы, секвенирование мРНК отдельных клеток показало низкую экспрессию ACE2 в легочных и бронхиальных тканях и, напротив, высокий уровень экспрессии в клетках почек и пищеварительной системы [5, 31]. Это свидетельствует о том, что помимо ACE2, в клетках существуют другие мишени SARS-CoV-2, посредством которых происходит инфицирование. В пользу такой точки зрения говорят также данные о наличии у переболевших COVID-19 нейтрализующих антител, которые связываются с SARS-CoV-2 по участку или участкам, отличным от RBD-домена [32, 33].
НЕЙРОПИЛИН
Установлено, что в поступлении коронавируса в клетки участвует мембранный белок нейропилин-1 (NRP1). Он работает как корецептор (host-factor) ACE2 при инфекции SARS-CoV-2 [34]. NRP1 – это гликопротеин с молекулярной массой около 130 кДа, выполняющий многообразные функции в нервной системе и других органах и тканях. Он был открыт как фактор, необходимый для формирования и направленного роста пучков нервных волокон [35]. Это происходит благодаря связыванию с NRP1 семафоринов класса 3, которые являются нейрональными хеморепеллентами [36]. Кроме того, установлена роль NRP1 в клеточной адгезии посредством гетерофильного молекулярного взаимодействия [37]. В эндотелиальных клетках NRP1 выступает как рецептор фактора роста эндотелия сосудов VEGF165, но не VEGF121, и регулирует ангиогенез [38]. Внеклеточная часть NRP1 (рис. 1) содержит домены, присутствующие в ряде белков, но образующие комбинацию, не характерную для других мембранных рецепторов [37]. В частности, а1/а2 имеют общие последовательности с компонентами комплемента C1r и C1s, b1/b2-подобные домены присутствуют в факторах свертывания крови V и VIII, домен МАМ (Meprin, A-5 protein, receptor protein-tyrosine phosphatase μ) содержится в металлоэндопептидазах мепринах и рецепторной протеинтирозинфосфатазе. Показано, что связывание семафоринов происходит с доменами a1/a2 и b1/b2, VEGF165 связывается с b1/b2, и они же обеспечивают клеточную адгезию [37].
Связывание VEGF165 и семафоринов 3 с доменом b1 осуществляется через имеющуюся у этих белков на С-конце многоосновную последовательность Arg/Lys-X-X-Arg/Lys (мотив CendR – C end Rule sequence). Установлено, что присоединение вирусной части SARS-CoV-2 к нейропилину происходит в этом же участке [34]. В случае SARS-CoV-2 фурин расщепляет белок S на белки S1 и S2, в результате чего на С-конце S1 экспонируется последовательность Arg-Arg-Ala-Arg (RRAR), с которой взаимодействует NRP1. Благодаря связыванию коронавируса с NRP1 облегчается проникновение вирусных частиц в клетки. Это происходит, по-видимому, в результате стабилизации С-концевой части (остатки 640–685) молекулы S1, что облегчает высвобождение N-концевого фрагмента (686–700 а.о.) субъединицы S2 и разделение субъединиц [39].
NRP1 инициирует инфицирование вирусом SARS-CoV-2 клеток обонятельного эпителия, несмотря на низкий уровень экспрессии в этих клетках ACE2 [40]. В опытах на макаках-резусах показано, что при инфицировании центральной нервной системы (ЦНС) вирусом SARS-CoV-2 реализуется механизм положительной обратной связи: в коре головного мозга происходит увеличение экспрессии NRP1, усиление иммунного ответа с высвобождением провоспалительных факторов и развитием вирусного энцефалита [41]. В варианте Омикрон рядом с мотивом CendR в S1 произошли мутации, благодаря которым связывание S1 c NRP1 энергетически более выгодно [42]. Возможно, это обуславливает повышенную инфекционность Омикрона. NRP1 рассматривается в качестве мишени для низкомолекулярных антиковидных препаратов, действующих по участку связывания CendR белка S1 [43].
CD147 (БАЗИГИН)
Трансмембранный гликопротеин базигин (от слов basic immunoglobulin, другие названия CD147 и EMMPRIN – extracellular matrix metalloproteinase inducer) относится к суперсемейству иммуноглобулинов. Он представлен двумя изоформами, различающимися по числу иммуноглобулиновых доменов в экстраклеточной части полипептидной цепи. У базигина-1 три домена, он локализован в ретине и служит рецептором трофического белка палочек сетчатки глаза (RdCVF), секретируемого колбочками [44]. У более распространенной изоформы – базигина-2, которая, собственно, и носит название базигин или CD147, имеется два иммуноглобулиновых домена [45]. CD147 выполняет функцию рецептора для циклофилинов, белка S100A9 и гликопротеина VI тромбоцитов. В иммуноглобулиновых доменах имеются гликозилированные участки, которыми CD147 связывается с лектинами – Е-селектином и галектином [45, 46].
CD147 (базигин) известен как белок, обеспечивающий поступление малярийного плазмодия в эритроциты [47]. Кроме этого, он выступает в качестве рецепторного белка при заражении клеток вирусом иммунодефицита человека типа 1, вирусами гепатита С и гепатита В, герпес-вирусом, ассоциированным с саркомой Капоши, вирусом SARS-CoV [48]. При инфицировании клеток ВИЧ-1 это происходит благодаря образованию тройного комплекса вируса с циклофилином А и CD147 [46].
Есть серия исследований, в которых получены данные об участии CD147 (базигина) в клеточной рецепции SARS-CoV-2. В работе большого коллектива китайских исследователей [49] было показано, что антитело против CD147 (базигина) меплазумаб подавляет амплификацию коронавируса в клетках Vero E6 и BEAS-2B. Напротив, экспрессия CD147 в невосприимчивых к SARS-CoV-2 клетках BHK-21 способствовала проникновению в них вируса. Меплазумаб подавлял заражение клеток Vero E6 исходным Уханьским вариантом SARS-CoV-2 и его последующими вариантами альфа, бета, гамма и дельта [50]. Подавление экспрессии CD147 в клетках CaLu3 (эпителиальные клетки человека из аденокарциномы легких HTB-55™) с помощью siRNA в 2–3 раза снижало инфицирование, которое определяли по синтезу мРНК SARS-CoV-2 [48]. Xu и соавт. [51] использовали псевдотипированный вирус SARS-CoV-2, выращенный в клетках с бессывороточной средой, и показали, что он может инфицировать клетки A549 с низким уровнем hACE2 при участии CD147. Кроме того, мыши hCD147KI-NSG, у которых был экспрессирован человеческий CD147, были более чувствительны к заражению коронавирусом по сравнению с однопометными мышами дикого типа WT-NSG [52]. CD147 относится к числу онкомаркеров, в связи с чем предполагается, что он может быть дополнительной важной мишенью при лечении COVID-19 у больных раком [53].
Несмотря на убедительные свидетельства в пользу важной роли CD147 в заражении новой коронавирусной инфекцией, остается неясным, как происходит взаимодействие вирусных частиц с CD147. Так, в исследованиях Shilts и соавт. [54] не было выявлено непосредственного связывания рекомбинантного пептида S1 и полного тримерного S-белка SARS-CoV-2 с CD147. В экспериментах исследователей из Великобритании и США [55] не обнаружено влияния поликлональных антител против CD147 на заражение клеток Vero E6 коронавирусом. Эти исследователи также не обнаружили прямого взаимодействия S-белка и RBD-домена с CD147. Возможно, что во взаимодействии CD147 и вирусных частиц участвуют дополнительные белки. Так, в заражении клеток SARS-CoV, как и в случае ВИЧ, участвует циклофилин А. Циклофилин А экспрессируется в цитоплазме, а также секретируется во внеклеточную среду. Однако в работе Fenizia и соавт. [48] показано, что, в отличие от SARS-CoV, комплекс CD147 с циклофилином А не участвует в заражении клеток коронавирусом SARS-CoV-2. По данным этих авторов, подавление экспрессии CD147 с помощью siRNA приводит к снижению уровня ACE2, и это рассматривается как один из возможных механизмов действия SARS-CoV-2, опосредованного CD147. В инфицированных псевдовирусом SARS-CoV-2 клетках Vero E6 и Huh-7 S-белок колокализуется с CD147 и Rab5a, что говорит о CD147-опосредованном эндоцитозе вирусных частиц [56]. CD147-опосредованный эндоцитоз SARS-CoV-2 происходит по Arf6-зависимому механизму.
ГЕПАРАН СУЛЬФАТ
Гепарансульфат (ГС) – это линейный полисахарид, структурной единицей которого являются сульфатированные D-глюкуроновая или L-идуроновая кислота, соединенная с сульфатированным N-ацетилглюкозамином. ГС присоединен к коровому, или стержневому, белку (core protein) и образует гепарансульфатные протеогликаны, экспрессированные на поверхности почти всех клеток млекопитающих и во внеклеточном матриксе. Имеется два типа мембранных гепарансульфатных протеогликанов – синдиканы и глипиканы [57]. Белок синдиканов связан с мембраной за счет трансмембранного С-концевого фрагмента, глипиканы ковалентно присоединены к фосфолипиду гликозилфосфатидилинозитолу (рис. 1). ГС выступает в качестве корецептора ACE2 при связывании вирусных частиц SARS-CoV-2 с клеточной мембраной. Clausen и соавт. показали [58], что ГС присоединяется к S-белку в участке домена RBD, расположенном рядом с участком связывания молекулы ACE2. По данным молекулярного докинга, при этом происходит образование тройного комплекса, причем гепарансульфат способствует разворачиванию сайта связывания ACE2. Сульфатированные полисахариды конкурентно ингибируют связывание S-белка с гепарансульфатом. Показано, что гепарин и эноксапарин (низкомолекулярный гепарин, мол. масса около 4500 Да) подавляют поступление псевдотипированного вируса SARS-CoV-2 в клетки HEK293T с IC50 соответственно 5.99 и 1.77 мкг/л [59].
CD209L (L-SIGN) и CD209/DC-SIGN
Гликопротеины CD209L, обозначаемые также L-SIGN (Liver/Lymph node-Specific ICAM-3 Grabbing Non-integrin) и CD209/DC-SIGN (Dendritic Cell-Specific Intеrcellular adhesion molecules (ICAM)-3 Grabbing Non-integrin), относятся к лектинам С-типа. Они непосредственно распознают широкий спектр вирусов HIV-1, Ebola, гепатита С, микобактерий Mycobacterium tuberculosis, паразитического микроорганизма Leishmania pifanoi и ряда других микроорганизмов (см. обзор [60]). Роль CD209L и CD209 в инфицировании клеток коронавирусами была первоначально установлена для SARS-CoV [61] и впоследствии подтверждена для SARS-CoV-2 [62, 63]. CD209L и CD209 выступают в качестве корецепторов ACE2 при связывании S-белка. В работе Thepaut и соавт. [64] показано, что лектин CD209L сам по себе при захвате вируса не вызывает заражение клеток Vero E6, не экспрессирующих ACE2, но способствует переносу вируса в клетки ACE2+ Vero E6. CD209L может выступать в качестве корецептора ACE2, образуя с ним гетеродимер [63]. Эти же авторы показали, что SARS-CoV-2 способен инфицировать эндотелиальные клетки (ЭК) сосудов человека, причем нокдаун CD209L или добавление растворимого CD209L снижает заражение. Вклад CD209L особенно выражен в клетках, в которых уровень экспрессии ACE2 низок, а экспрессия CD209L, напротив, высока. Это относится к ЭК синуса печени человека и эндотелиальным клеткам лимфатических узлов [30, 65]. Методом конфокальной микроскопии вирусные частицы выявлены в ЭК синуса печени в аутопсийном материале от больных COVID [65]. ЭК синуса печени являются основными источниками фактора Виллебранда (vWF) и фактора свертывания VIII (FVIII). Заражение этих ЭК коронавирусом, по-видимому, является одной из причин повышенного свертывания крови и тромбообразования при коронавирусной инфекции. В миелоидных клетках SARS-CoV-2 усиливает провоспалительные реакции посредством CD209 и других лектинов С-типа – секрецию интерейкинов 1b и 8, цитокинов CCL2/3, хемокина CXLC10 [66]. Специфические антагонисты CD209L PolyMan26 и другие гликомиметики маннозида подавляют связывание S-белка и инфицирование клеток [67]. Есть данные об участии CD209L и других лектинов C-типа в активации коронавирусом SARS-CoV-2 секреции миелоидными клетками провоспалительных цитокинов и хемокинов [66].
GRP78 (GLUCOSE-REGULATED PROTEIN 78)
GRP78 (Glucose-Regulated Protein 78) является белком теплового шока – шапероном, в нормальных клетках осуществляющим рефолдинг белков в эндоплазматическом ретикулуме либо направляющим развернутые белки в клеточные системы деградации [68]. При стрессе эндоплазматического ретикулума происходит гиперэкспрессия и транслокация GRP78 на клеточную мембрану [69], где он также проводит обработку большого числа развернутых белков. Однако экспонированный на плазматической мембране GRP78, помимо восстановительной работы по рефолдингу белков, способствует проникновению в клетку патогенов – вирусов, бактерий и грибков. Гиперэкспрессия GRP78 наблюдается на мембранах различных раковых клеток, что повышает агрессивность онкологического заболевания. Молекулярный докинг выявил предполагаемое взаимодействие между GRP78 и рецептор-связывающим доменом (RBD) S-белка SARS-CoV-2 [70]. GRP78 может образовывать комплекс с SARS-2-S и ACE2 на поверхности и в околоядерной области, характерной для эндоплазматического ретикулума в клетках VeroE6-ACE2, причем показано, что субстрат-связывающий домен GRP78 имеет решающее значение для этого взаимодействия [71]. Чтобы исследовать роль GRP78, авторами цитированной работы был проведен нокдаун GRP78 в клетках VeroE6-ACE2. Потеря GRP78 заметно снижала экспрессию ACE2 на клеточной поверхности. Был сделан вывод, что GRP78 представляет собой вспомогательный фактор для проникновения SARS-CoV-2 в клетки [71].
РЕЦЕПТОРНАЯ ТИРОЗИНКИНАЗА AXL
По данным Wang и соавт. [5], рецепторная тирозинкиназа AXL специфически взаимодействует с N-концевым доменом S1-субъединицы S-белка коронавируса SARS-CoV-2. Избыточная экспрессия AXL в клетках HEK293T способствовала проникновению SARS-CoV-2 так же эффективно, как и избыточная экспрессия ACE2. В клетках легких H1299 и первичных эпителиальных клетках легких человека, где уровень ACE2 низкий, а уровень AXL высок, нокаут последней значительно снижал инфицирование SARS-CoV-2. Растворимая человеческая рекомбинантная тирозинкиназа AXL блокировала инфекцию SARS-CoV-2 в клетках, экспрессирующих высокие уровни AXL. Авторы статьи показали, что уровень экспрессии AXL хорошо коррелирует с уровнем S-белка SARS-CoV-2 в клетках жидкости бронхоальвеолярного лаважа у пациентов с COVID-19. На основе этих данных был сделан вывод о том, что AXL является новым рецептором для SARS-CoV-2, который может играть важную роль в распространении коронавирусной инфекции дыхательной системы человека. Лигандом AXL является белок GAS6, служащий мостиком, соединяющим фосфатидилсерин вирусной мембраны с AXL [72]. Таким способом клетки инфицируют ряд вирусов, в частности Денге, Западного Нила и Эбола. В статье [5] авторы на основании собственных данных утверждают, что в случае SARS-CoV-2 работает некий альтернативный механизм.
ASGR1 и KREMEN1
Недавно были выявлены два новых рецептора, специфично связывающих S-белок SARS-CoV-2 [73]. Этими белками оказались ASGR1 (asialoglycoprotein receptor-1) и KREMEN1 (Kringle Containing Transmembrane Protein 1). Для выявления рецепторов был проведен скрининг 5054 мембранных белков человека (91.6% от общего количества предполагаемых белков мембран) на предмет связывания ими S-белка. Каждый из этих белков по отдельности экспрессировали в клетках HEK293E с нокаутом ACE2 и определяли связывание с данными клетками экстраклеточного фрагмента S-белка вирусов SARS-CoV-2, SARS-CoV и MERS. В результате было обнаружено 12 белков, способных специфически связывать с высоким сродством S-белок SARS-CoV-2 (Kдис< 525 нМ), включая уже известные рецепторы ACE2 и CD209L (L-SIGN). Константы диссоциации Kдис S-белка SARS-CoV-2 с ASGR1 и KREMEN1 составили 94.8 и 19.3 нМ, что сопоставимо с Kдис для ACE2 – 12.4 нМ. Авторы показали, что KREMEN1 связывается с доменами RBD, NTD и S2-доменом экстраклеточной части S-белка с наибольшей аффинностью по отношению к RBD, а ASGR1 эффективно связывается с доменом RBD и с меньшим сродством с доменом NTD. S-белки вирусов SARS-CoV и MERS не связывались c ASGR1 и KREMEN1.
Было исследовано, экспрессия каких белков из числа выявленных рецепторов экстраклеточного фрагмента S-белка коронавируса делает клетки, не экспрессирующие ACE2, чувствительными к заражению вирусом SARS-CoV-2. Оказалась, что ACE2-независимое инфицирование происходит только с ASGR1 и KREMEN1. Этот результат не противоречит ранее полученным данным о том, что CD209L (L-SIGN) участвует в транспортировке коронавируса в клетки в паре с ACE2 [64]. Возможно, другие связывающие S-белок мембранные белки из приведенного в работе [73] списка, включая родственный CD209L другой лектин С-типа, CD207, также работают как функциональные рецепторы коронавируса в связке с ACE2. Несмотря на то, что ASGR1 и KREMEN1 могут работать как независимые транспортеры SARS-CoV-2, при их совместной экспрессии с ACE2 способность SARS-CoV-2 инфицировать клетки синергично увеличивалась [73].
Структуры ASGR1 и KREMEN1 схематично представлены на рис. 1. В экстраклеточной части KREMEN1 имеется три домена – Krm (Kringle domain), WSC и CUB [74]. Главную роль в связывании S-белка играют домен CUB в KREMEN1. В ASGR1 связывание осуществляется с доменом лектина С-типа. KREMEN1 в организме выполняет функции высокоаффинного рецептора DKK1 – антагониста Wnt-сигнального пути [74, 75] и, кроме того, является переносчиком в клетки некоторых энтеровирусов [76]. ASGR1 экспрессируется преимущественно в гепатоцитах и регулирует концентрацию гликопротеинов в плазме крови, осуществляя эндоцитоз десиалированных гликопротеинов [77]. Известна его роль как рецептора вируса гепатита С [78].
СКЭВЕНДЖЕР-РЕЦЕПТОР ЛИПОПРОТЕИНОВ ВЫСОКОЙ ПЛОТНОСТИ В ТИПА 1
В работе Wei и соавт. [79] показано, что скэвенджер-рецептор (рецептор-мусорщик) липопротеинов высокой плотности (ЛПВП) B типа 1 (SR-B1) способствует зависимому от ACE2 проникновению SARS-CoV-2. SR-B1 облегчает проникновение SARS-CoV-2 в клетки благодаря более прочному прикреплению вируса к мембране. Авторами показано, что S-белок способен связывать холестерин и ЛПВП, что, возможно, обеспечивает присоединение коронавируса к рецепторам SR-B1. Известна другая патогенетическая роль SR-B1 – он переносит в гепатоциты вирусы гепатита С и вируса Денге [80]. Рецептор SR-B1 в наибольшей степени экспрессирован в печени и в стероидогенных клетках, где осуществляет поступление холестерина соответственно для синтеза желчных кислот и стероидных гормонов [81].
ТЯЖЕЛАЯ ЦЕПЬ НЕМЫШЕЧНОГО МИОЗИНА IIA (NONMUSCLE MYOSIN HEAVY CHAIN IIA)
Тяжелая цепь немышечного миозина IIA (НТЦМ IIA) была идентифицирована как еще один белок, способствующий инфицированию клеток легких человека коронавирусом SARS-CoV-2 [82]. Было показано, что NTG-домен S1‑субъединицы и субъединица S2 непосредственно взаимодействуют с С-концевым доменом тяжелой цепи. Удаление генетическим путем НТЦМ IIA значительно снижало псевдовирусную инфекцию SARS-CoV-2 в клетках дикого типа (WT) A549 и Calu-3, а сверхэкспрессия НТЦМ IIA ее усиливала.
ЗАКЛЮЧЕНИЕ
Интенсивное изучение коронавируса SARS-CoV-2 показало, что механизмы его действия на клетки человека более разнообразные и сложные, чем это исходно представлялось. Получены данные о наличии у клеток дополнительных помимо ACE2 рецепторов для S-белка, расширены представления о протеолитическом праймировании коронавируса, появились новые сведения о его влиянии на сигнальные системы клетки. Изучение этих вопросов имеет принципиальное значение, поскольку у людей разного возраста, в частности, взрослых и детей, у людей с разными сопутствующими заболеваниями течение новой коронавирусной инфекции проявляет существенные особенности, которые могут быть обусловлены характером экспрессии и активности тех или иных рецепторов коронавируса SARS-CoV-2 и вспомогательных белков, участвующих в заражении клеток. Изучение молекулярных механизмов взаимодействия вирусов с клетками должно также способствовать пониманию того, почему уже появившиеся и возможные будущие штаммы коронавируса различаются по клеточной и органной специфичности и по тяжести воздействия на организм человека.
Конфликт интересов. Авторы декларируют отсутствие явных и потенциальных конфликтов интересов, связанных с публикацией настоящей статьи.
Источники финансирования. Работа выполнена при поддержке Российского научного фонда (грант № 21-15-00441).
Соответствие принципам этики. Настоящая статья не содержит описания каких-либо исследований с участием людей или животных в качестве объектов.
Список литературы
Peeri N.C., Shrestha N., Rahman M.S., Zaki R., Tan Z., Bibi S., Baghbanzadeh M., Aghamohammadi N., Zhang W., Haque U. 2020. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: What lessons have we learned? Int. J. Epidemiol. 49, 717–726. https://doi.org/10.1093/ije/dyaa033
Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 181, 271–280 e278. https://doi.org/10.1016/j.cell.2020.02.052
Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. 2020. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 117, 11 727–11 734. https://doi.org/10.1073/pnas.2003138117
Bayati A., Kumar R., Francis V., McPherson P.S. 2021. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 296, 100306. https://doi.org/10.1016/j.jbc.2021.100306
Wang S., Qiu Z., Hou Y., Deng X., Xu W., Zheng T., Wu P., Xie S., Bian W., Zhang C., Sun Z., Liu K., Shan C., Lin A., Jiang S., Xie Y., Zhou Q., Lu L., Huang J., Li X. 2021. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 31, 126–140. https://doi.org/10.1038/s41422-020-00460-y
Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. 2003. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J. Virol. 77, 8801–8811. https://doi.org/10.1128/jvi.77.16.8801-8811.2003
Schibli D.J., Weissenhorn W. 2004. Class I and class II viral fusion protein structures reveal similar principles in membrane fusion. Mol. Membr. Biol. 21, 361–371. https://doi.org/10.1080/09687860400017784
Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. 2020. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 41, 1141–1149. https://doi.org/10.1038/s41401-020-0485-4
Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 367, 1260–1263. https://doi.org/10.1126/science.abb2507
Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Jr., Rawson S., Rits-Volloch S., Chen B. 2020. Distinct conformational states of SARS-CoV-2 spike protein. Science. 369, 1586–1592. https://doi.org/10.1126/science.abd4251
Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 181, 281–292 e286. https://doi.org/10.1016/j.cell.2020.02.058
Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. 2020. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral. Res. 176, 104742. https://doi.org/10.1016/j.antiviral.2020.104742
Zhang Q., Xiang R., Huo S., Zhou Y., Jiang S., Wang Q., Yu F. 2021. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct. Target Ther. 6, 233. https://doi.org/10.1038/s41392-021-00653-w
Guney C., Akar F. 2021. Epithelial and endothelial expressions of ACE2: SARS-CoV-2 entry routes. J. Pharm. Pharm. Sci. 24, 84–93. https://doi.org/10.18433/jpps31455
Zhao M.M., Yang W.L., Yang F.Y., Zhang L., Huang W.J., Hou W., Fan C.F., Jin R.H., Feng Y.M., Wang Y.C., Yang J.K. 2021. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct. Target Ther. 6, 134. https://doi.org/10.1038/s41392-021-00558-8
Zhao M.M., Zhu Y., Zhang L., Zhong G., Tai L., Liu S., Yin G., Lu J., He Q., Li M.J., Zhao R.X., Wang H., Huang W., Fan C., Shuai L., Wen Z., Wang C., He X., Chen Q., Liu B., Xiong X., Bu Z., Wang Y., Sun F., Yang J.K. 2022. Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies. Cell Discov. 8, 53. https://doi.org/10.1038/s41421-022-00419-w
Padmanabhan P., Desikan R., Dixit N.M. 2020. Targeting TMPRSS2 and Cathepsin B/L together may be synergistic against SARS-CoV-2 infection. PLoS Comput. Biol. 16, e1008461. https://doi.org/10.1371/journal.pcbi.1008461
Prasad K., AlOmar S.Y., Almuqri E.A., Rudayni H.A., Kumar V. 2021. Genomics-guided identification of potential modulators of SARS-CoV-2 entry proteases, TMPRSS2 and Cathepsins B/L. PLoS One. 16, e0256141. https://doi.org/10.1371/journal.pone.0256141
Prasad K., Ahamad S., Kanipakam H., Gupta D., Kumar V. 2021. Simultaneous Inhibition of SARS-CoV-2 Entry Pathways by Cyclosporine. ACS Chem. Neurosci. 12, 930–944. https://doi.org/10.1021/acschemneuro.1c00019
Evans J.P., Liu S.L. 2021. Role of host factors in SARS-CoV-2 entry. J. Biol. Chem. 297, 100847. https://doi.org/10.1016/j.jbc.2021.100847
Matsuyama S., Ujike M., Morikawa S., Tashiro M., Taguchi F. 2005. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. USA. 102, 12543–12547. https://doi.org/10.1073/pnas.0503203102
Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 426, 450–454. https://doi.org/10.1038/nature02145
Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. 2005. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 11, 875–879. https://doi.org/10.1038/nm1267
Lin C.R., Bahmed K., Simborio H., Hayek H., Bolla S., Marchetti N., Criner G.J., Kosmider B. 2021. Expression of SARS-CoV-2 entry factors in human alveolar type II cells in aging and emphysema. Biomedicines. 9. https://doi.org/10.3390/biomedicines9070779
Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 579, 270–273. https://doi.org/10.1038/s41586-020-2012-7
Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. 2020. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 367, 1444–1448. https://doi.org/10.1126/science.abb2762
Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., Breitbart R.E., Acton S. 2000. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ. Res. 87, E1-9. https://doi.org/10.1161/01.res.87.5.e1
Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. 2000. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 275, 33 238–33 243. https://doi.org/10.1074/jbc.M002615200
Hikmet F., Mear L., Edvinsson A., Micke P., Uhlen M., Lindskog C. 2020. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 16, e9610. https://doi.org/10.15252/msb.20209610
Li J., Gao J., Xu Y.P., Zhou T.L., Jin Y.Y., Lou J.N. 2007. Expression of severe acute respiratory syndrome coronavirus receptors, ACE2 and CD209L in different organ derived microvascular endothelial cells. Zhonghua Yi Xue Za Zhi. 87, 833–837.
Han X., Zhou Z., Fei L., Sun H., Wang R., Chen Y., Chen H., Wang J., Tang H., Ge W., Zhou Y., Ye F., Jiang M., Wu J., Xiao Y., Jia X., Zhang T., Ma X., Zhang Q., Bai X., Lai S., Yu C., Zhu L., Lin R., Gao Y., Wang M., Wu Y., Zhang J., Zhan R., Zhu S., Hu H., Wang C., Chen M., Huang H., Liang T., Chen J., Wang W., Zhang D., Guo G. 2020. Construction of a human cell landscape at single-cell level. Nature. 581, 303–309. https://doi.org/10.1038/s41586-020-2157-4
Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., Chen Z., Guo Y., Zhang J., Li Y., Song X., Chen Y., Xia L., Fu L., Hou L., Xu J., Yu C., Li J., Zhou Q., Chen W. 2020. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 369, 650–655. https://doi.org/10.1126/science.abc6952
Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., Bentlage A.E.H., van Haaren M.M., Guerra D., Burger J.A., Schermer E.E., Verheul K.D., van der Velde N., van der Kooi A., van Schooten J., van Breemen M.J., Bijl T.P.L., Sliepen K., Aartse A., Derking R., Bontjer I., Kootstra N.A., Wiersinga W.J., Vidarsson G., Haagmans B.L., Ward A.B., de Bree G.J., Sanders R.W., van Gils M.J. 2020. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 369, 643–650. https://doi.org/10.1126/science.abc5902
Daly J.L., Simonetti B., Klein K., Chen K.E., Williamson M.K., Anton-Plagaro C., Shoemark D.K., Simon-Gracia L., Bauer M., Hollandi R., Greber U.F., Horvath P., Sessions R.B., Helenius A., Hiscox J.A., Teesalu T., Matthews D.A., Davidson A.D., Collins B.M., Cullen P.J., Yamauchi Y. 2020. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 370, 861–865. https://doi.org/10.1126/science.abd3072
Fujisawa H., Kitsukawa T., Kawakami A., Takagi S., Shimizu M., Hirata T. 1997. Roles of a neuronal cell-surface molecule, neuropilin, in nerve fiber fasciculation and guidance. Cell Tissue Res. 290, 465–470. https://doi.org/10.1007/s004410050954
Telley L., Cadilhac C., Cioni J.M., Saywell V., Jahannault-Talignani C., Huettl R.E., Sarrailh-Faivre C., Dayer A., Huber A.B., Ango F. 2016. Dual function of NRP1 in axon guidance and subcellular target recognition in cerebellum. Neuron. 91, 1276–1291. https://doi.org/10.1016/j.neuron.2016.08.015
Shimizu M., Murakami Y., Suto F., Fujisawa H. 2000. Determination of cell adhesion sites of neuropilin-1. J. Cell Biol. 148, 1283–1293. https://doi.org/10.1083/jcb.148.6.1283
Soker S., Takashima S., Miao H.Q., Neufeld G., Klagsbrun M. 1998. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell. 92, 735–745. https://doi.org/10.1016/s0092-8674(00)81402-6
Li Z.L., Buck M. 2021. Neuropilin-1 assists SARS-CoV-2 infection by stimulating the separation of Spike protein S1 and S2. Biophys. J. 120, 2828–2837. https://doi.org/10.1016/j.bpj.2021.05.026
Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., Smura T., Levanov L., Szirovicza L., Tobi A., Kallio-Kokko H., Osterlund P., Joensuu M., Meunier F.A., Butcher S.J., Winkler M.S., Mollenhauer B., Helenius A., Gokce O., Teesalu T., Hepojoki J., Vapalahti O., Stadelmann C., Balistreri G., Simons M. 2020. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 370, 856–860. https://doi.org/10.1126/science.abd2985
Gao C.C., Li M., Deng W., Ma C.H., Chen Y.S., Sun Y.Q., Du T., Liu Q.L., Li W.J., Zhang B., Sun L., Liu S.M., Li F., Qi F., Qu Y., Ge X., Liu J., Wang P., Niu Y., Liang Z., Zhao Y.L., Huang B., Peng X.Z., Yang Y., Qin C., Tong W.M., Yang Y.G. 2022. Differential transcriptomic landscapes of multiple organs from SARS-CoV-2 early infected rhesus macaques. Protein Cell. https://doi.org/10.1007/s13238-022-00915-5
Baindara P., Roy D., Mandal S.M., Schrum A.G. 2022. Conservation and enhanced binding of SARS-CoV-2 Omicron spike protein to coreceptor neuropilin-1 predicted by docking analysis. Infect. Dis. Rep. 14, 243–249. https://doi.org/10.3390/idr14020029
Kolaric A., Jukic M., Bren U. 2022. Novel small-molecule inhibitors of the SARS-CoV-2 spike protein binding to neuropilin 1. Pharmaceuticals (Basel). 15 (2), 165. https://doi.org/10.3390/ph15020165
Ait-Ali N., Fridlich R., Millet-Puel G., Clerin E., Delalande F., Jaillard C., Blond F., Perrocheau L., Reichman S., Byrne L.C., Olivier-Bandini A., Bellalou J., Moyse E., Bouillaud F., Nicol X., Dalkara D., van Dorsselaer A., Sahel J.A., Leveillard T. 2015. Rod-derived cone viability factor promotes cone survival by stimulating aerobic glycolysis. Cell. 161, 817–832. https://doi.org/10.1016/j.cell.2015.03.023
Muramatsu T. 2016. Basigin (CD147), a multifunctional transmembrane glycoprotein with various binding partners. J. Biochem. 159, 481–490. https://doi.org/10.1093/jb/mvv127
Pushkarsky T., Zybarth G., Dubrovsky L., Yurchenko V., Tang H., Guo H., Toole B., Sherry B., Bukrinsky M. 2001. CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proc. Natl. Acad. Sci. USA. 98, 6360–6365. https://doi.org/10.1073/pnas.111583198
Crosnier C., Bustamante L.Y., Bartholdson S.J., Bei A.K., Theron M., Uchikawa M., Mboup S., Ndir O., Kwiatkowski D.P., Duraisingh M.T., Rayner J.C., Wright G.J. 2011. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature. 480, 534–537. https://doi.org/10.1038/nature10606
Fenizia C., Galbiati S., Vanetti C., Vago R., Clerici M., Tacchetti C., Daniele T. 2021. SARS-CoV-2 entry: At the crossroads of CD147 and ACE2. Cells. 10 (6), 1434. https://doi.org/10.3390/cells10061434
Wang K., Chen W., Zhang Z., Deng Y., Lian J.Q., Du P., Wei D., Zhang Y., Sun X.X., Gong L., Yang X., He L., Zhang L., Yang Z., Geng J.J., Chen R., Zhang H., Wang B., Zhu Y.M., Nan G., Jiang J.L., Li L., Wu J., Lin P., Huang W., Xie L., Zheng Z.H., Zhang K., Miao J.L., Cui H.Y., Huang M., Zhang J., Fu L., Yang X.M., Zhao Z., Sun S., Gu H., Wang Z., Wang C.F., Lu Y., Liu Y.Y., Wang Q.Y., Bian H., Zhu P., Chen Z.N. 2020. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target Ther. 5, 283. https://doi.org/10.1038/s41392-020-00426-x
Geng J., Chen L., Yuan Y., Wang K., Wang Y., Qin C., Wu G., Chen R., Zhang Z., Wei D., Du P., Zhang J., Lin P., Zhang K., Deng Y., Xu K., Liu J., Sun X., Guo T., Yang X., Wu J., Jiang J., Li L., Zhang K., Wang Z., Zhang J., Yan Q., Zhu H., Zheng Z., Miao J., Fu X., Yang F., Chen X., Tang H., Zhang Y., Shi Y., Zhu Y., Pei Z., Huo F., Liang X., Wang Y., Wang Q., Xie W., Li Y., Shi M., Bian H., Zhu P., Chen Z.N. 2021. CD147 antibody specifically and effectively inhibits infection and cytokine storm of SARS-CoV-2 and its variants delta, alpha, beta, and gamma. Signal Transduct. Target Ther. 6, 347. https://doi.org/10.1038/s41392-021-00760-8
Xu C., Wang A., Geng K., Honnen W., Wang X., Bruiners N., Singh S., Ferrara F., D’Angelo S., Bradbury A.R.M., Gennaro M.L., Liu D., Pinter A., Chang T.L. 2021. Human immunodeficiency viruses pseudotyped with SARS-CoV-2 spike proteins infect a broad spectrum of human cell lines through multiple entry mechanisms. Viruses. 13. https://doi.org/10.3390/v13060953
Badeti S., Jiang Q., Naghizadeh A., Tseng H.C., Bushkin Y., Marras S.A.E., Nisa A., Tyagi S., Chen F., Romanienko P., Yehia G., Evans D., Lopez-Gonzalez M., Alland D., Russo R., Gause W., Shi L., Liu D. 2022. Development of a novel human CD147 knock-in NSG mouse model to test SARS-CoV-2 viral infection. Res. Sq. rs.3.rs-1431484. https://doi.org/10.21203/rs.3.rs-1431484/v1
Xia P., Dubrovska A. 2020. Tumor markers as an entry for SARS-CoV-2 infection? FEBS J. 287 (17), 3677–3680. https://doi.org/10.1111/febs.15499
Shilts J., Crozier T.W.M., Greenwood E.J.D., Lehner P.J., Wright G.J. 2021. No evidence for basigin/CD147 as a direct SARS-CoV-2 spike binding receptor. Sci. Rep. 11, 413. https://doi.org/10.1038/s41598-020-80464-1
Ragotte R.J., Pulido D., Donnellan F.R., Hill M.L., Gorini G., Davies H., Brun J., McHugh K., King L.D.W., Skinner K., Miura K., Long C.A., Zitzmann N., Draper S.J. 2021. Human basigin (CD147) does not directly interact with SARS-CoV-2 spike glycoprotein. mSphere. 6, e0064721. https://doi.org/10.1128/mSphere.00647-21
Zhou Y.Q., Wang K., Wang X.Y., Cui H.Y., Zhao Y., Zhu P., Chen Z.N. 2022. SARS-CoV-2 pseudovirus enters the host cells through spike protein-CD147 in an Arf6-dependent manner. Emerg. Microbes Infect. 11, 1135–1144. https://doi.org/10.1080/22221751.2022.2059403
De Pasquale V., Quiccione M.S., Tafuri S., Avallone L., Pavone L.M. 2021. Heparan sulfate proteoglycans in viral infection and treatment: A special focus on SARS-CoV-2. Int. J. Mol. Sci. 22. https://doi.org/10.3390/ijms22126574
Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N., Thacker B.E., Glass C.A., Yang Z., Torres J.L., Golden G.J., Bartels P.L., Porell R.N., Garretson A.F., Laubach L., Feldman J., Yin X., Pu Y., Hauser B.M., Caradonna T.M., Kellman B.P., Martino C., Gordts P., Chanda S.K., Schmidt A.G., Godula K., Leibel S.L., Jose J., Corbett K.D., Ward A.B., Carlin A.F., Esko J.D. 2020. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 183, 1043–1057. https://doi.org/10.1016/j.cell.2020.09.033
Tandon R., Sharp J.S., Zhang F., Pomin V.H., Ashpole N.M., Mitra D., McCandless M.G., Jin W., Liu H., Sharma P., Linhardt R.J. 2021. Effective inhibition of SARS-CoV-2 entry by heparin and enoxaparin derivatives. J. Virol. 95. https://doi.org/10.1128/JVI.01987-20
Khoo U.S., Chan K.Y., Chan V.S., Lin C.L. 2008. DC-SIGN and L-SIGN: The SIGNs for infection. J. Mol. Med. (Berl.). 86, 861–874. https://doi.org/10.1007/s00109-008-0350-2
Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J., Thomas W.D., Jr., Thackray L.B., Young M.D., Mason R.J., Ambrosino D.M., Wentworth D.E., Demartini J.C., Holmes K.V. 2004. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 101, 15748–15753. https://doi.org/10.1073/pnas.0403812101
Amraie R., Napoleon M.A., Yin W., Berrigan J., Suder E., Zhao G., Olejnik J., Gummuluru S., Muhlberger E., Chitalia V., Rahimi N. 2020. CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2 and are differentially expressed in lung and kidney epithelial and endothelial cells. bioRxiv. https://doi.org/10.1101/2020.06.22.165803
Amraei R., Yin W., Napoleon M.A., Suder E.L., Berrigan J., Zhao Q., Olejnik J., Chandler K.B., Xia C., Feldman J., Hauser B.M., Caradonna T.M., Schmidt A.G., Gummuluru S., Muhlberger E., Chitalia V., Costello C.E., Rahimi N. 2021. CD209L/L-SIGN and CD209/DC-SIGN Act as Receptors for SARS-CoV-2. ACS Cent Sci. 7, 1156–1165. https://doi.org/10.1021/acscentsci.0c01537
Thepaut M., Luczkowiak J., Vives C., Labiod N., Bally I., Lasala F., Grimoire Y., Fenel D., Sattin S., Thielens N., Schoehn G., Bernardi A., Delgado R., Fieschi F. 2021. DC/L-SIGN recognition of spike glycoprotein promotes SARS-CoV-2 trans-infection and can be inhibited by a glycomimetic antagonist. PLoS Pathog. 17, e1009576. https://doi.org/10.1371/journal.ppat.1009576
Kondo Y., Larabee J.L., Gao L., Shi H., Shao B., Hoover C.M., McDaniel J.M., Ho Y.C., Silasi-Mansat R., Archer-Hartmann S.A., Azadi P., Srinivasan R.S., Rezaie A.R., Borczuk A., Laurence J.C., Lupu F., Ahamed J., McEver R.P., Papin J.F., Yu Z., Xia L. 2021. L-SIGN is a receptor on liver sinusoidal endothelial cells for SARS-CoV-2 virus. JCI Insight. 6. https://doi.org/10.1172/jci.insight.148999
Lu Q., Liu J., Zhao S., Gomez Castro M.F., Laurent-Rolle M., Dong J., Ran X., Damani-Yokota P., Tang H., Karakousi T., Son J., Kaczmarek M.E., Zhang Z., Yeung S.T., McCune B.T., Chen R.E., Tang F., Ren X., Chen X., Hsu J.C.C., Teplova M., Huang B., Deng H., Long Z., Mudianto T., Jin S., Lin P., Du J., Zang R., Su T.T., Herrera A., Zhou M., Yan R., Cui J., Zhu J., Zhou Q., Wang T., Ma J., Koralov S.B., Zhang Z., Aifantis I., Segal L.N., Diamond M.S., Khanna K.M., Stapleford K.A., Cresswell P., Liu Y., Ding S., Xie Q., Wang J. 2021. SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and Tweety family member 2. Immunity. 54, 1304–1319 e1309. https://doi.org/10.1016/j.immuni.2021.05.006
Pollastri S., Delaunay C., Thepaut M., Fieschi F., Bernardi A. 2022. Glycomimetic ligands block the interaction of SARS-CoV-2 spike protein with C-type lectin co-receptors. Chem. Commun. (Camb). 58, 5136–5139. https://doi.org/10.1039/d2cc00121g
Ibrahim I.M., Abdelmalek D.H., Elfiky A.A. 2019. GRP78: A cell’s response to stress. Life Sci. 226, 156–163. https://doi.org/10.1016/j.lfs.2019.04.022
Elfiky A.A., Baghdady A.M., Ali S.A., Ahmed M.I. 2020. GRP78 targeting: Hitting two birds with a stone. Life Sci. 260, 118317. https://doi.org/10.1016/j.lfs.2020.118317
Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. 2020. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 80, 554–562. https://doi.org/10.1016/j.jinf.2020.02.026
Carlos A.J., Ha D.P., Yeh D.W., Van Krieken R., Tseng C.C., Zhang P., Gill P., Machida K., Lee A.S. 2021. The chaperone GRP78 is a host auxiliary factor for SARS-CoV-2 and GRP78 depleting antibody blocks viral entry and infection. J. Biol. Chem. 296, 100759. https://doi.org/10.1016/j.jbc.2021.100759
Morizono K., Chen I.S. 2014. Role of phosphatidylserine receptors in enveloped virus infection. J. Virol. 88, 4275–4290. https://doi.org/10.1128/JVI.03287-13
Gu Y., Cao J., Zhang X., Gao H., Wang Y., Wang J., He J., Jiang X., Zhang J., Shen G., Yang J., Zheng X., Hu G., Zhu Y., Du S., Zhu Y., Zhang R., Xu J., Lan F., Qu D., Xu G., Zhao Y., Gao D., Xie Y., Luo M., Lu Z. 2022. Receptome profiling identifies KREMEN1 and ASGR1 as alternative functional receptors of SARS-CoV-2. Cell Res. 32, 24–37. https://doi.org/10.1038/s41422-021-00595-6
Nakamura T., Nakamura T., Matsumoto K. 2008. The functions and possible significance of Kremen as the gatekeeper of Wnt signalling in development and pathology. J. Cell Mol. Med. 12, 391–408.
Mao B., Wu W., Davidson G., Marhold J., Li M., Mechler B.M., Delius H., Hoppe D., Stannek P., Walter C., Glinka A., Niehrs C. 2002. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature. 417, 664–667. https://doi.org/10.1038/nature756
Staring J., van den Hengel L.G., Raaben M., Blomen V.A., Carette J.E., Brummelkamp T.R. 2018. KREMEN1 Is a Host Entry Receptor for a Major Group of Enteroviruses. Cell Host Microbe. 23, 636–643 e635. https://doi.org/10.1016/j.chom.2018.03.019
Hoober J.K. 2020. ASGR1 and its enigmatic relative, CLEC10A. Int. J. Mol. Sci. 21. 48818. https://doi.org/10.3390/ijms21144818
Saunier B., Triyatni M., Ulianich L., Maruvada P., Yen P., Kohn L.D. 2003. Role of the asialoglycoprotein receptor in binding and entry of hepatitis C virus structural proteins in cultured human hepatocytes. J. Virol. 77, 546–559. https://doi.org/10.1128/jvi.77.1.546-559.2003
Wei C., Wan L., Yan Q., Wang X., Zhang J., Yang X., Zhang Y., Fan C., Li D., Deng Y., Sun J., Gong J., Yang X., Wang Y., Wang X., Li J., Yang H., Li H., Zhang Z., Wang R., Du P., Zong Y., Yin F., Zhang W., Wang N., Peng Y., Lin H., Feng J., Qin C., Chen W., Gao Q., Zhang R., Cao Y., Zhong H. 2020. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat. Metab. 2, 1391–1400. https://doi.org/10.1038/s42255-020-00324-0
Kluck G.E.G., Yoo J.A., Sakarya E.H., Trigatti B.L. 2021. Good Cholesterol Gone Bad? HDL and COVID-19. Int. J. Mol. Sci. 22. https://doi.org/10.3390/ijms221910182
Shen W.J., Asthana S., Kraemer F.B., Azhar S. 2018. Scavenger receptor B type 1: expression, molecular regulation, and cholesterol transport function. J. Lipid Res. 59, 1114–1131. https://doi.org/10.1194/jlr.R083121
Chen J., Fan J., Chen Z., Zhang M., Peng H., Liu J., Ding L., Liu M., Zhao C., Zhao P., Zhang S., Zhang X., Xu J. 2021. Nonmuscle myosin heavy chain IIA facilitates SARS-CoV-2 infection in human pulmonary cells. Proc. Natl. Acad. Sci. USA. 118. https://doi.org/10.1073/pnas.2111011118
Дополнительные материалы отсутствуют.
Инструменты
Биологические мембраны: Журнал мембранной и клеточной биологии


