Биология моря, 2022, T. 48, № 3, стр. 201-211
Ортологи генов семейства миогенных регуляторных факторов (MRF) и их возможные функции у иглокожих
В. А. Нижниченко 1, *, И. Ю. Долматов 1
1 Национальный научный центр морской биологии им. А.В. Жирмунского ДВО РАН
690041 Владивосток, Россия
* E-mail: 0hm@mail.ru
Поступила в редакцию 05.08.2021
После доработки 03.12.2021
Принята к публикации 27.01.2022
- EDN: QKOUOR
- DOI: 10.31857/S0134347522030068
Аннотация
Гены семейства MRF, играющие важную роль в развитии мышечной системы, есть у всех животных. Исследование доступных транскриптомов и геномов разных представителей Echinodermata показало наличие у этих животных ортологов гена MyoD позвоночных. У морских лилий и морских ежей присутствуют три гена: MyoD1, MyoD2 и MyoD3; морские звезды, по-видимому, утратили ген MyoD3, а голотурии – MyoD2. При регенерации мышц у голотурии Eupentacta fraudatrix значительно увеличивалась экспрессия гена MyoD3, в то время как число транскриптов MyoD1 не изменялось. Это может указывать на участие MyoD3 в регуляции миогенеза у голотурий.
Гены семейства MRF (myogenic regulatory factors) играют ключевую роль в миогенезе позвоночных (Buckingham, Rigby, 2014). Семейство состоит из четырех генов: MyoD, Myf5, Myogenin (MyoG) и MRF4. Предполагается, что у последнего общего предка всех Chordata имелся только один ген этого семейства (Dehal et al., 2003). У беспозвоночных к настоящему времени обнаружены ортологи только одного представителя MRF – гена MyoD (Venuti et al., 1991; Araki et al., 1994; Adams et al., 2000; Howard-Ashby et al., 2006). Вероятно, эволюция MRF у вторичноротых животных происходила независимо, как у Chordata, так и у Ambulacraria.
Гены семейства MRF кодируют транскрипционные факторы, относящиеся к классу II basic helix-loop-helix (bHLH). Они играют важную роль в развитии мышц. Их функция заключается в регуляции пролиферации, выводе клеток-предшественников из клеточного цикла, в дифференциации и формировании фенотипа миоцита скелетных мышц. Все гены семейства MRF имеют сходное строение и включают участки, кодирующие N-концевой трансактивационный домен с участком, богатым цистеином/гистидином (N-TAD), центральный участок с мотивом bHLH, включающим два домена – basic myogenic и helix-loop-helix (HLH), а также С-концевой трансактивационный домен Myf5, который содержит Helix III (Singh, Dilworth, 2013). Домены HLH и basic myogenic отвечают за гетеродимеризацию с E-протеинами – белками другого семейства bHLH. Образующийся комплекс способен связываться с последовательностью CANNTG в регуляторных участках целевых генов (Lassar et al., 1991). N- и С-концевые трансактивационные домены важны для дополнительной регуляции функций транскрипционных факторов MRF (Weintraub et al., 1991; Bergstrom, Tapscott, 2001). В отличие от высококонсервативных доменов HLH и basic myogenic, N- и C-концевые последовательности достаточно вариабельны и различаются у белков разных семейств MRF (Ishibashi et al., 2005).
Высокая гомология участков bHLH всех членов семейства MRF указывает на их функциональную схожесть, в то время как различия в других доменах обуславливают наличие у отдельных белков MRF специализированных функций (Atchley et al., 1994). Например, MYF5 и MyoD не только запускают транскрипцию, но могут создавать открытую структуру хроматина в генах, связанных с миогенезом. В свою очередь, это позволяет белку MyoG регулировать транскрипцию этих генов (Lassar et al., 1991). Помимо участия в развитии мышц белки семейства MRF играют ключевую роль в их регенерации (Grounds et al., 1992; Zhou, Bornemann, 2001; Pavlath et al., 2003; Karalaki et al., 2009; White et al., 2016).
Иглокожие – одна из групп беспозвоночных, показывающих хорошие способности к регенерации. После значительных повреждений у них могут восстанавливаться как внешние придатки, так и внутренние органы (Candia Carnevali, 2006; Долматов и др., 2014). Восстановление у этих животных происходит без участия стволовых клеток за счет дедифференцировки сохранившихся дифференцированных клеток (Mladenov et al., 1989; García-Arrarás et al., 1998; Dolmatov, Ginanova, 2001; Mashanov et al., 2005, 2008; Долматов, Машанов, 2007; García-Arrarás, Dolmatov, 2010). В частности, показано, что у голотурий при повреждении продольной мышечной ленты (ПМЛ) новые миоциты формируются в результате миогенной дифференцировки клеток целомического эпителия, покрывающего мышцу (Dolmatov et al., 1996; Dolmatov, Ginanova, 2001). Высокая скорость и полнота восстановления ПМЛ после поперечной перерезки делают голотурий удобным модельным объектом для изучения регенерации мышечной системы у иглокожих (Dolmatov, Ginanova, 2001; García-Arrarás, Dolmatov, 2010). Особенности миогенеза у этих животных достаточно хорошо изучены на клеточном уровне как в развитии, так и при регенерации (Dolmatov, Ivantey, 1993; Dolmatov et al., 1996; Ginanova, 1998; Dolmatov, Ginanova, 2001; Dolmatov, 2010). Однако отсутствуют данные о молекулярных механизмах и об участии в этом процессе генов MRF у иглокожих.
В настоящее время наличие ортологов гена MyoD показано только для двух видов морских ежей. У Strongylocentrotus purpuratus (Stimpson, 1857) найдены гены MyoD1, MyoD2 и MyoD3 (Howard-Ashby et al., 2006), а у Lytechinus variegatus (Lamarck, 1816) – ген MyoD1 (Venuti et al., 1991). По другим классам иглокожих и генам семейства данные отсутствуют. Нет информации и об участии генов MRF в миогенезе у Echinodermata. В связи с этим нами предпринята попытка выявить гены данного семейства у разных видов иглокожих, а также определить их гомологию и участие в регенерации.
МАТЕРИАЛ И МЕТОДИКА
Для анализа использовали имеющиеся в базах данных SPbase (http://www.echinobase.org/Echinobase) и NCBI (https://www.ncbi.nlm.nih.gov) геномы морских ежей Strongylocentrotus purpuratus (PRJNA13728), Eucidaris tribuloides (Lamarck, 1816) (PRJNA63057) и Lytechinus variegatus (PRJNA657258); морских звезд Patiria miniata (Brandt, 1835) (PRJNA683060) и Acanthaster planci (Linnaeus, 1758) (PRJNA397419); голотурий Apostichopus japonicus (Selenka, 1867) (PRJNA354676) и A. parvimensis (Clark, 1913) (PRJNA182998) и морской лилии Anneissia japonica (Müller, 1841) (PRJNA615663).
Использовали также транскриптомы кишки морской лилии Himerometra robustipinna (Carpenter, 1881) (Долматов и др., 2014) и голотурии Eupentacta fraudatrix (D’yakonov, Baranova & Savel’eva, 1958) (Boyko et al., 2020), амбулакров и целомического эпителия голотурий Cladolabes schmeltzii (Ludwig, 1875) (Dolmatov et al., 2018) и Holothuria glaberrima (Risso, 1826) (PRJNA158511), а также яйцеклеток голотурии Sclerodactyla briareus (Lesueur, 1824) (PRJNA236087) и морской звезды Henricia sp. (PRJNA236087). Поиск выполняли в программе Ugene и в базе NCBI (https://www.ncbi.nlm.nih.gov) алгоритмами blastp, tblastx, blastn.
Для определения рамок считывания использовали blastx. Окончательную сборку контигов, определение и выравнивание предполагаемых аминокислотных последовательностей проводили в программе Mega X с помощью алгоритма Muscle. Последовательности, использованные для выравнивания, доступны по https://drive.google.com/drive/folders/1VIoilLfYZSiti-9lXkPPIGG9-5f2ut7g? usp=sharing. Сравнение предполагаемых аминокислотных последовательностей выполняли инструментом blastp (align two sequences).
Для построения филогенетических деревьев использовали кодирующие последовательности транскриптов, предполагаемые аминокислотные последовательности и консервативные участки предполагаемых аминокислотных последовательностей представителей четырех классов иглокожих: голотурий A. japonicus, A. parvimensis, S. bri-areus, E. fraudatrix, C. schmeltzii и H. glaberrima, морских ежей S. purpuratus, E. tribuloides и L. variegatus, морских звезд P. miniata, Henricia sp. и A. planci и морских лилий H. robustipinna и A. japonica с добавлением последовательностей насекомых (Drosophila melanogaster), полухордовых (Saccoglossus kowalevskii) и хордовых (Branchiostoma belcheri, Ciona intestinalis, Danio rerio, Xenopus laevis, Mus musculus). Для более достоверного определения принадлежности последовательностей генов и белков иглокожих к MRF в анализ добавлены нуклеотидные и предполагаемые аминокислотные последовательности других членов суперсемейства bHLH (NeuroD1, NeuroG1, bHLH21, dimmed, twist, bHLH15, mesp, bHLH23, atonal, nato) хордовых (B. belcheri) и полухордовых (S. kowalevski).
Для построения филогенетических деревьев использовали два алгоритма: MrBayes (Guindon et al., 2010) и RAxML-HPC BlackBox (8.2.12) онлайн сервиса CIPRES (http://www.phylo.org). Для построения филогенетических деревьев алгоритмом MrBayes нуклеотидные и аминокислотные последовательности анализировали с помощью Partitionfinder 2.1.1 (Ronquist et al., 2012; Lanfear et al., 2016). Визуализацию деревьев осуществляли в программе FigTree.
Полимеразная цепная реакция в реальном времени (кПЦР)
В качестве объекта исследования использовали взрослых особей голотурии E. fraudatrix в норме и через 10 и 20 сут после поперечного разрезания амбулакра (стенки тела и ПМЛ). Суммарную РНК выделяли с помощью TRI Reagent (MRC).
Для проведения кПЦР использовали по 3 особи животных на каждый срок регенерации и норму. Для синтеза кДНК брали 1 мкг тотальной РНК, набор MMLV (Evrogen) согласно рекомендациям изготовителя и Random праймеры. В качестве референсных генов использовали Tubulin (Tub) и Elongated Factor 1α (EF1α). Реакцию проводили с использованием набора SYBER Green RT-PCR kit (Синтол, Россия) на приборе С1000 Thermal Cycler CFX96 Real-Time System (Bio-Rad, США). Реакционная смесь (20 мкл) состояла из 9.8 мкл mQ, 2 мкл 10 × PCR буфера Б (Синтол), 2 мкл 2.5 мМ dNTP (Синтол), 1 мкл 25 мМ MgCl2 (Синтол), 0.2 мкл Syn Taq ДНК-полимеразы (Синтол) и по 2 мкл 5 мМ праймеров: на Tub – TubβF2 и TubβR2; на EF1α – EF1αF2 и EF1αR2; на MyoD3 голотурии E. fraudatrix – MyoD3F1 и MyoD3R1; на MyoD1 голотурии E. fraudatrix – MyoDF1 и MyoDR1. Последовательности использованных праймеров приведены в таблице 1. Программа амплификации: этап 1 – 1 мин при температуре 95°C, этап 2 –10 с при 96°C, этап 3 – 30 с при 63°C, этап 4 – 10 с при 72°C. Количество циклов – 40. Каждую пробу амплифицировали дважды для предупреждения ошибок. Уровень базовой линии для определения, на каких циклах должны выходить гены, был установлен на 100. Результаты обработаны с использованием программ Bio-Rad CFX Manager 2.1 и Microsoft Excel, ezANOVA.
Таблица 1.
Нуклеотидные последовательности праймеров для проведения количественной ПЦР
| Название праймера | Последовательность (5'–3') |
|---|---|
| TubβF2 | CAGCCGTCTGTGACATTCC |
| TubβR2 | TCAGTAAACTCCATCTAATCCAT |
| EF1αF2 | ACATCAACATTGTGGTCATCGG |
| EF1αR2 | ACGCTCAGCCTTCAGTTTGTCT |
| MyoD3F1 | ACCCATCATCGGTAAGAGAAACAGG |
| MyoD3R1 | GTTCGTCCATCTTCACCACTCCTG |
| MyoD1F1 | GGCGGCTCAGAAAGGTCAAC |
| MyoD1R1 | CGTCTGTCATTCCGTCTGATGTATT |
РЕЗУЛЬТАТЫ
У всех исследованных видов иглокожих не обнаружены гомологи генов семейств Myf, MyoG и MRF, но найдены гомологи MyoD. У разных видов иглокожих число ортологов MyoD различалось и варьировало от 1 до 3.
Анализ геномов морской звезды A. planci, голотурии A. japonicus и морской лилии A. japonica показал наличие у этих видов гена MyoD1. Кодируемый им белок содержал 4 домена: N-TAD, basic myogenic, HLH и Myf5. В геномах морской звезды P. miniata, морского ежа E. tribuloides и голотурии A. parvimensis ген MyoD1 кодировал белок, который содержал только домены N-TAD, basic myogenic и HLH.
Полные транскрипты MyoD1 найдены в транскриптомах голотурий E. fraudatrix и H. glaberrima, а также морской лилии H. robustipinna. При этом транскрипты E. fraudatrix и H. glaberrima содержали участки, кодирующие все четыре домена белка MyoD1, а у H. robustipinna Myf5 отсутствовал. В транскриптоме голотурии C. schmeltzii обнаружен неполный транскрипт MyoD1, имеющий участки, кодирующие все четыре домена. В транскриптомах голотурии S. briareus и морской звезды Henricia sp. продукты этого гена не отмечены (табл. 2).
Таблица 2.
Наличие гена MyoD1 и доменная структура MyoD1 у представителей разных классов иглокожих
| Класс | Вид | Домен | |||
|---|---|---|---|---|---|
| N-TAD | Basic myogenic | HLH | Myf5 | ||
| Голотурии | Apostichopus japonicus | + | + | + | + |
| Cladolabes schmeltzii | + | + | + | + | |
| Eupentacta fraudatrix | + | + | + | + | |
| Holothuria glaberrima | + | + | + | + | |
| Apostichopus parvimensis | + | + | + | – | |
| Sclerodactyla briareus | Ген не обнаружен | ||||
| Морские ежи | Eucidaris tribuloides | + | + | + | – |
| Lytechinus variegatus | + | + | + | + | |
| Strongylocentrotus purpuratus | + | + | + | + | |
| Морские звезды | Acanthaster planci | + | + | + | + |
| Henricia sp. | Ген не обнаружен | ||||
| Patiria miniata | + | + | + | – | |
| Морские лилии | Anneissia japonica | + | + | + | + |
| Himerometra robustipinna | + | + | + | – | |
В геномах морских ежей L. variegatus и E. tribuloides, морской звезды A. planci и морской лилии A. japonica обнаружен ген MyoD2, кодирующий белок с доменами N-TAD, basic myogenic и HLH. В геномах морской звезды P. miniata, а также голотурий A. parvimensis и A. japonicus этот ген отсутствовал. Дополнительно в транскриптоме морской звезды Henricia sp. выявлен полный транскрипт MyoD2. Он содержал участки, кодирующие домены N-TAD, basic myogenic и HLH. В транскриптомах остальных исследованных видов иглокожих продукты MyoD2 отсутствовали (табл. 3).
Таблица 3.
Наличие гена MyoD2 и доменная структура MyoD2 у представителей разных классов иглокожих
| Класс | Вид | Домен | |||
|---|---|---|---|---|---|
| N-TAD | Basic myogenic | HLH | Myf5 | ||
| Голотурии | Apostichopus japonicus | Ген не обнаружен | |||
| Cladolabes schmeltzii | Ген не обнаружен | ||||
| Eupentacta fraudatrix | Ген не обнаружен | ||||
| Holothuria glaberrima | Ген не обнаружен | ||||
| Apostichopus parvimensis | Ген не обнаружен | ||||
| Sclerodactyla briareus | Ген не обнаружен | ||||
| Морские ежи | Eucidaris tribuloides | + | + | + | – |
| Lytechinus variegatus | + | + | + | – | |
| Strongylocentrotus purpuratus | + | + | + | – | |
| Морские звезды | Acanthaster planci | + | + | + | – |
| Henricia sp. | + | + | + | – | |
| Patiria miniata | Ген не обнаружен | ||||
| Морские лилии | Anneissia japonica | + | + | + | – |
| Himerometra robustipinna | Ген не обнаружен | ||||
Анализ геномов морского ежа L. variegatus, голотурии A. japonicus и морской лилии A. japonica показал наличие у этих животных гена MyoD3. Кодируемая им аминокислотная последовательность содержала консервативные участки basic myogenic и HLH. В транскриптомах морской лилии H. robustipinna и голотурии S. briareus выявлены полные транскрипты гена MyoD3. В каждом из них содержался участок, кодирующий домены basic myogenic и HLH. В транскриптоме голотурии E. fraudatrix найден неполный транскрипт MyoD3, который, тем не менее, содержал области, соответствующие basic myogenic и HLH. У остальных исследованных иглокожих ген MyoD3 или его продукты не обнаружены (рис. 1, табл. 4). Домены basic myogenic MyoD3 в нуклеотидной и предполагаемой аминокислотной последовательностях значительно отличались от таковых MyoD1 у позвоночных и иглокожих, а также MyoD2 у иглокожих (рис. 1).
Рис. 1.
Домены basic myogenic и HLH в транскрипционных факторах MyoD у позвоночных, иглокожих и Drosophila melanogaster.
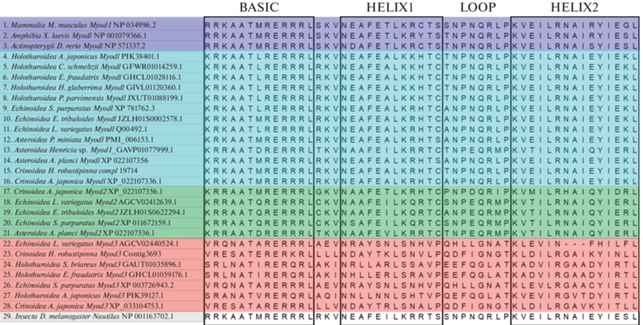
Таблица 4.
Наличие гена MyoD3 и доменная структура MyoD3 у представителей разных классов иглокожих
| Класс | Вид | Домен | |||
|---|---|---|---|---|---|
| N-TAD | Basic myogenic | HLH | Myf5 | ||
| Голотурии | Apostichopus japonicus | – | + | + | – |
| Cladolabes schmeltzii | Ген не обнаружен | ||||
| Eupentacta fraudatrix | – | + | + | – | |
| Holothuria glaberrima | Ген не обнаружен | ||||
| Apostichopus parvimensis | Ген не обнаружен | ||||
| Sclerodactyla briareus | – | + | + | – | |
| Морские ежи | Eucidaris tribuloides | Ген не обнаружен | |||
| Lytechinus variegatus | – | + | + | – | |
| Strongylocentrotus purpuratus | – | + | + | – | |
| Морские звезды | Acanthaster planci | Ген не обнаружен | |||
| Henricia sp. | Ген не обнаружен | ||||
| Patiria miniata | Ген не обнаружен | ||||
| Морские лилии | Anneissia japonica | – | + | + | – |
| Himerometra robustipinna | – | + | + | – | |
На филогенетических деревьях, построенных методами MrBayes и RAxML-HPC BlackBox, уровни поддержки (MrBayes posterior probabilities и bootstrap support levels, соответственно) в ключевых для данного исследования узлах чуть выше при первом алгоритме. Сила поддержки в большей степени зависит от типа анализируемых последовательностей: построения по аминокислотным последовательностям дают уровни поддержки выше (как MrBayes posterior probabilities, так и bootstrap support levels), чем по нуклеотидным. Однако топология деревьев в целом остается неизменной при использовании как разных типов последовательностей, так и разных алгоритмов. В качестве примера приведено филогенетическое дерево, построенное по аминокислотным последовательностям методом MrBayes (рис. 2).
Рис. 2.
Филогенетическое дерево дивергенции генов семейства MRF иглокожих и хордовых. Дерево сгенерировано с помощью метода MrBayes с использованием предполагаемых аминокислотных последовательностей белков. Значения в узлах дерева отображают Байесовские апостериорные вероятности (Bayesian posterior probabilities).
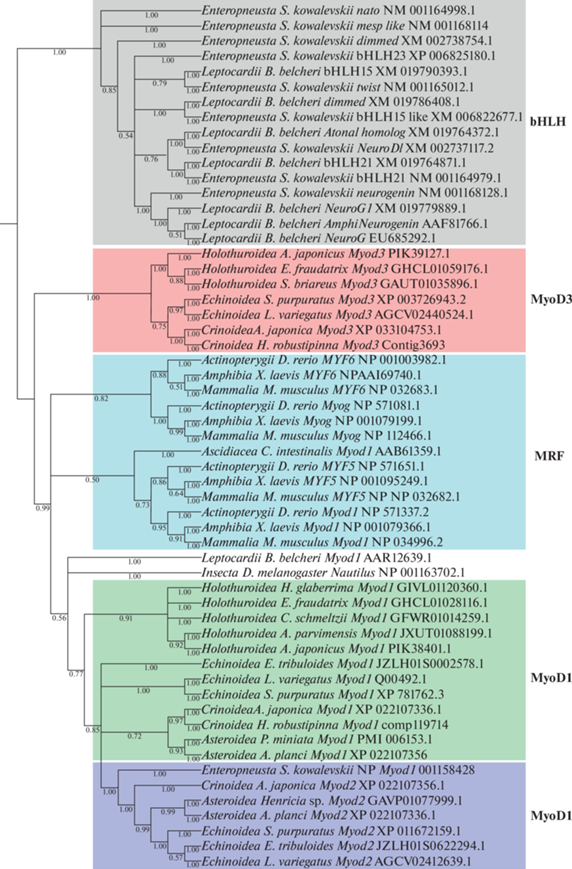
Изучаемые гены и кодируемые ими белки по-разному группируются на филогенетических деревьях относительно транскрипционных факторов bHLH. Прежде всего, независимо от способа построения, последовательности MyoD1 и MyoD3 иглокожих достоверно разделяются. На построенных филогенетических деревьях гены MyoD1 и кодируемые ими белки исследованных иглокожих образуют общую группу с членами семейства MRF хордовых. При этом выявленные последовательности MyoD2 иглокожих входят в кластер MyoD1; MyoD3 образуют обособленную группу, которая, тем не менее, имеет связь с генами семейства MRF (рис. 2). Все выявленные последовательности иглокожих и последовательности семейства MRF хордовых животных достоверно отделены от остальных транскрипционных факторов bHLH (рис. 2).
Анализ гомологии предполагаемых аминокислотных последовательностей изучаемых генов подтвердил результаты филогении. Наибольшее сходство выявлено между видами иглокожих по соответствующим генам: Myod1 – 72%, MyoD2 – 71% и MyoD3 – 53%. Гомология предполагаемых аминокислотных последовательностей транскриптов MyoD1 исследованных иглокожих с транскрипционными факторами семейства MRF и остальными членами bHLH позвоночных составила 56 и 35.7% соответственно. Приблизительно такой же уровень сходства показали и последовательности MyoD2. Гомология с белками MRF позвоночных составила 53%, а с другими белками bHLH – 32%. Наибольшие отличия характеризуют MyoD3 иглокожих, сходство которых с белками семейства MRF составило 40%, а с остальными членами bHLH – 34%.
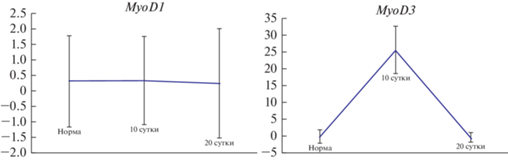
Исследование динамики экспрессии генов MyoD1 и MyoD3 при восстановлении продольной мышечной ленты у голотурии E. fraudatrix, проведенное для выявления роли генов MRF в регенерации иглокожих, показало, что эти гены различаются по уровням экспрессии как у интактных животных, так и на разных стадиях миогенеза (рис. 3). В норме референсные гены Tub и EF1α в среднем по трем запускам выходили на циклах 21 и 19 соответственно. В неповрежденных тканях ген MyoD1 выходил в среднем на 26, а MyoD3 – только на 32 цикле. При регенерации уровень экспрессии MyoD1 не менялся и оставался на уровне нормы. Количество транскриптов MyoD3 через 10 сут после повреждения увеличивалось в 25 раз, а через 20 сут возвращалось к значениям, близким к норме.
ОБСУЖДЕНИЕ
Исследование показало, что ген MyoD1 присутствует у всех изученных иглокожих, за исключением голотурии S. briareus и морской звезды Henricia sp. Причиной отсутствия гена MyoD1 могла быть его утрата. Однако, если учесть, что у остальных изученных иглокожих ген MyoD1 присутствует и играет важную роль в поддержке скелетогенной линии как в процессе эмбрионального развития, так и в постнатальном периоде (Andrikou et al., 2013), то утрата этого консервативного гена представляется маловероятной. Другим объяснением может быть неполнота данных в транскриптомах S. briareus и Henricia sp. У этих видов анализировали только мРНК, находившуюся в яйцеклетках. Вероятно, ген MyoD1 не относится к генам материнского эффекта и его транскрипты не накапливаются в половых клетках.
На построенных филогенетических деревьях гены MyoD1 и кодируемые ими белки исследованных иглокожих образуют общую группу с членами семейства MRF хордовых и стоят максимально далеко от прочих транскрипционных факторов bHLH, что говорит об их явной принадлежности к MRF (рис. 2).
Данные по имеющимся геномам и транскриптомам показали, что у всех изученных иглокожих ген MyoD1 кодирует четыре домена. Такую же доменную структуру имеют и ортологи MyoD1 исследованных видов полухордовых и хордовых. В то же время наши исследования показали, что у морской звезды P. miniata, морского ежа E. tribuloides и голотурии A. parvimensis MyoD1 не содержит С-концевой трансактивационный домен Myf5.
Нормальное развитие мышечной системы в отсутствие домена Myf5 или даже одного из транскрипционных факторов можно объяснить тем, что члены семейства MRF имеют сходные функции (Baugh, Hunter, 2006). Ранее показано, что у позвоночных при нокауте одного из генов этого семейства мышцы развиваются нормально (Rudnicki et al., 1992), однако при инактивации двух генов миогенез не происходит, что указывает на частичное дублирование функций этих генов (Rudnicki et al., 1993). У беспозвоночных функции транскрипционных факторов, регулирующих развитие мышц, также перекрываются (Fukushige et al., 2007). У нематоды Caenorhabditis elegans гены hlh-1, unc-120 и hnd-1 (ортологи MyoD1, MADS-box/SRF и HAND/bHLH-1 позвоночных соответственно) сообща регулируют развитие мышц в эмбриогенезе, однако утрата любого из них мало влияет на миогенез (Baugh, Hunter, 2006). Показано, что ген nautilus – ортолог MyoD1 у дрозофилы D. melanogaster также не является необходимым для эмбрионального миогенеза (Balagopalan et al., 2001). Весьма примечательно, что hlh-1 и nautilus тоже не имеют Myf5 (Michelson et al., 1990). Вероятно, отсутствие трансактивационного домена в MyoD1 у иглокожих P. miniata, E. tribuloides и A. parvimensis может компенсироваться работой других генов сети регуляции миогенеза.
Нормальное развитие мускулатуры у этих животных может достигаться не только за счет отличия доменной структуры транскрипционных факторов, но и благодаря их активации на разных этапах морфогенетических процессов. В частности, у мышей Myf5 и MyoD1 сохранили сходную специфичность сайта связывания, но кодирующие их гены экспрессируются на разных этапах спецификации и дифференцировки клеток скелетных мышц. Включаясь в работу первым, Myf5 модифицирует хроматин в сайтах связывания Myf5, однако не взаимодействует с Pol II и не активирует транскрипцию генов, тогда как MyoD1 связывается с теми же сайтами, но активно рекрутирует Pol II и активирует транскрипцию генов (Conerly et al., 2016). Подобный механизм, где управление морфогенетическими процессами осуществляется за счет темпоральной последовательности активизации MyoD1 и других транскрипционных факторов, может присутствовать у P. miniata и E. tribuloides. Этим можно объяснить отсутствие нарушений в развитии при утрате трансактивационного домена.
Среди всех изученных иглокожих лишь у морской звезды Henricia sp. в транскриптоме присутствовал MyoD2. Найденный в геномах морских ежей L. variegatus и E. tribuloides, морской звезды A. planci и морской лилии A. japonica ген MyoD2 содержал участки, кодирующие домены N-TAD, basic myogenic и HLH. У голотурий данный ген не найден. Возможно, голотурии утратили его, однако это предположение нуждается в проверке на большем числе видов.
На построенных филогенетических деревьях последовательности MyoD2 иглокожих составляют отдельную группу, входящую в один кластер вместе с MyoD1 иглокожих и хордовых, а также с MyoD1, Myf5, Myog, Myf6 позвоночных, что говорит о явной принадлежности MyoD2 к семейству MRF. Последовательность морской лилии A. japonica, ранее обозначенная как SUM-1-like (XP_033102045.1), на построенных деревьях вошла в группу MyoD2, в то время как последовательность XP_033101843.1, также обозначенная как SUM-1-like, вошла в группу MyoD1 (рис. 2).
Ген MyoD3 и его транскрипты выявлены у голотурий, морских ежей и морских лилий. В то же время у всех исследованных видов морских звезд он отсутствовал, что, скорее всего, говорит о его утрате. Домен basic myogenic в MyoD3 иглокожих сильно отличается от такового в MyoD1 и MyoD2 иглокожих и позвоночных главным образом заменой первой аминокислоты: лизина (MyoD1 и MyoD2 иглокожих) или аргинина (MyoD1 позвоночных) на серин или валин. Лизин и аргинин – оснóвные аминокислоты, они взаимозаменяемы для нормальной работы домена, в то время как валин (нейтральная гидрофобная аминокислота) и серин (нейтральная полярная аминокислота) отличаются по физико-химическим свойствам от первых двух и различаются между собой (рис. 1). Данные различия ставят под вопрос работоспособность этого домена в MyoD3. В связи с чем MyoD3 не имеет возможности связываться с ДНК, поскольку именно домен basic myogenic отвечает за эту функцию (Lassar et al., 1991). Такие bHLH белки с нерабочим basic myogenic доменом относятся к группе d, члены которой выступают антагонистами bHLH белков группы a, в которую входят MyoD1 и MyoD2 (Van Doren et al., 1991, 1992). Необходимы дальнейшие исследования, чтобы выяснить, каким образом MyoD3 участвует в транскрипционных взаимодействиях.
На построенных филогенетических деревьях последовательности MyoD3 стоят отдельно от MRF, но все же образуют с ними одну большую обособленную от остальных транскрипционных факторов группу (рис. 2). Филогенетический анализ, сравнение предполагаемых аминокислотных последовательностей и результаты поиска через BLAST позволяют сделать вывод, что гены MyoD3, хотя и относятся к семейству MRF, все же имеют слабую гомологию с его представителями. Низкая гомология внутри группы MyoD3 и столь большие различия в доменах наводят на мысль, что эти уникальные для иглокожих гены сильно дивергировали после дупликации.
Исследование показало различия в экспрессии и, вероятно, в функциях генов семейства MRF у голотурий. При регенерации мышц у E. fraudatrix экспрессия гена MyoD3 увеличивалась значительно, в то время как число транскриптов гена MyoD1 не изменялось. Это может указывать на особую роль MyoD3 в регенерации мышцы и стенки тела у E. fraudatrix. Вероятно, ген MyoD3 у голотурий может быть одним из регуляторов миогенеза. В связи с этим необходимы дальнейшие исследования его функций не только при регенерации, но и в ходе личиночного развития.
Список литературы
Долматов И.Ю., Машанов В.С. Регенерация у голотурий. Владивосток: Дальнаука. 2007. 212 с.
Долматов И.Ю., Бобровская Н.В., Гирич А.С. Иглокожие как модельные объекты для изучения механизмов регенерации // Вестн. СПбГУ. 2014. Сер. 3. Вып. 3. С. 96–112.
Adams M., Celniker S., Holt R. et al. The genome sequence of Drosophila melanogaster // Science. 2000. V. 287. P. 2185–2195.
Andrikou C., Iovene E., Rizzo F. et al. Myogenesis in the sea urchin embryo: the molecular fingerprint of the myoblast precursors // EvoDevo. 2013. V. 4. Art. № 33.
Araki S., Saiga H., Makabe K., Satoh N. Expression of AMD 1, a gene for a MyoD 1-related factor in the ascidian Halocynthia roretzi // Roux’s Arch. Dev. Biol. 1994. V. 203. P. 320–327.
Atchley W., Fitch W., Bronner-Fraser M. Molecular evolution of the MyoD family of transcription factors // Proc. Nat. Acad. Sci. U.S.A. 1994. V. 91. P. 11522–11526.
Balagopalan L., Keller C., Abmayr S. Loss-of-function mutations reveal that the Drosophila nautilus gene is not essential for embryonic myogenesis or viability // Dev. Biol. 2001. V. 231. P. 374–382.
Baugh L., Hunter C. MyoD, modularity, and myogenesis: conservation of regulators and redundancy in C. elegans // Genes Dev. 2006. V. 20. P. 3342–3346.
Bergstrom D., Tapscott S. Molecular distinction between specification and differentiation in the myogenic basic helix-loop-helix transcription factor family // Mol. Cell. Biol. 2001. V. 21. № 7. P. 2404–2412.
Boyko A., Girich A., Tkacheva E., Dolmatov I. The Eupentacta fraudatrix transcriptome provides insights into regulation of cell transdifferentiation // Sci. Rep. 2020. V. 10. Art. № 1522.
Buckingham M., Rigby P. Gene regulatory networks and transcriptional mechanisms that control myogenesis // Dev. Cell. 2014. V. 28 P. 225–238.
Candia Carnevali M.D. Regeneration in Echinoderms: repair, regrowth, cloning // Invertebr. Surviv. J. 2006. V. 3. P. 64–76.
Conerly M., Yao Z., Zhong J. et al. Distinct activities of Myf5 and MyoD indicate separate roles in skeletal muscle lineage specification and differentiation // Dev. Cell. 2016. V. 36. P. 375–385.
Dehal P., Satou Y., Campbell R. et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins // Science. 2003. V. 298. P. 2157–2167.
Dolmatov I. Development and evolution of the muscle system in the Echinodermata // Echinoderms: Durham – Proceedings of the 12th International Echinoderm Conference. 2010. P. 163–166.
Dolmatov I., Afanasyev S., Boyko A. Molecular mechanisms of fission in echinoderms: transcriptome analysis // PLoS One. 2018.V. 13. Art. ID e0195836.
Dolmatov I., Eliseikina M., Bulgakov T. et al. Muscle rege-neration in the holothurian Stichopus japonicus // Roux’s Arch. Dev. Biol. 1996. V. 205. P. 486–493.
Dolmatov I., Ginanova T. Muscle regeneration in holothurians // Microsc. Res. 2001. V. 55. P. 452–463.
Dolmatov I., Ivantey V. Histogenesis of longitudinal muscle bands in holothurians // RJDB. 1993. V. 24. P. 67–72.
Fukushige T., Brodigan T., Schriefer L. et al. Defining the transcriptional redundancy of early bodywall muscle development in C. elegans: evidence for a unified theory of animal muscle development // Genes Dev. 2007. V. 20. P. 3395–3406.
García-Arrarás J., Dolmatov I. Echinoderms: potential model systems for studies on muscle regeneration // Curr. Pharm. Des. 2010. V. 16. P. 942–955.
García-Arrarás J., Estrada-Rodgers L., Santiago R. et al. Cellular mechanisms of intestine regeneration in the sea cucumber, Holothuria glaberrima Selenka (Holothuroidea: Echinodermata) // J. Exp. Zool. 1998. V. 281 P. 288–304.
Ginanova T. DNA synthesis during muscle regeneration in sea cucumber // Biol. Bull. 1998. V. 25. P. 9–13.
Grounds M., Garrett K., Lai M. et al. Identification of skeletal muscle precursor cells in vivo by use of MyoD1 and myogenin probes // Cell Tissue Res. 1992. V. 267. P. 99–104.
Guindon S., Dufayard J., Lefort V. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0 // Syst. Biol. 2010. V. 59. P. 307–321.
Howard-Ashby M., Materna S., Brown C. et al. Gene families encoding transcription factors expressed in early development of Strongylocentrotus purpuratus // Dev. Biol. 2006. V. 300. P. 90–107.
Ishibashi J., Perry R., Asakura A., Rudnicki M. MyoD induces myogenic differentiation through cooperation of its NH2- and COOH-terminal regions // J. Cell Biol. 2005. V. 171. P. 471–482.
Karalaki M., Fili S., Philippou A., Kontsilieris M. Muscle regeneration: cellular and molecular events // In Vivo. 2009. V. 23. P. 779–796.
Lanfear R., Frandsen P., Wright A. et al. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses // Mol. Biol. Evol. 2016. V. 34. № 3. P. 772–773.
Lassar A., Davis R., Wright W. et al. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo // Cell. 1991. V. 66. P. 305–315.
Mashanov V., Dolmatov I., Heinzeller T. Transdifferentiation in holothurian gut regeneration // Biol. Bull. 2005. V. 209. № 3. P. 184–193.
Mashanov V., Zueva O., Heinzeller T. Regeneration of the radial nerve cord in a holothurian: a promising new model system for studying post-traumatic recovery in the adult nervous system // Tissue Cell. 2008. V. 40. P. 351–372.
Michelson A., Abmayr S., Bate M. et al. Expression of a MyoD family member prefigures muscle pattern in Drosophila embryos // Genes Dev. 1990. V. 4. P. 2086–2097.
Mladenov P., Igdoura S., Asotra S. et al. Purification and partial characterization of an autotomy-promoting factor from the sea star Pycnopodia helianthoides // Biol. Bull. 1989. V. 176. P. 169–175.
Pavlath G., Dominov J., Kegley K. et al. Regeneration of transgenic skeletal muscles with altered timing of expression of the basic helix-loop-helix muscle regulatory factor MRF4 // Am. J. Pathol. 2003. V. 162. P. 1685–1691.
Ronquist F., Teslenko M., van der Mark P. et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space // Syst. Biol. 2012. V. 61. P. 539–542.
Rudnicki M., Braun T., Hinuma S., Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development // Cell. 1992. V. 71. P. 383–390.
Rudnicki M., Schnegelsberg P., Stead R. et al. MyoD or Myf-5 is required for the formation of skeletal muscle // Cell. 1993. V. 75. P. 1351–1359.
Singh K., Dilworth F. Differential modulation of cell cycle progression distinguishes members of the myogenic regulatory factor family of transcription factors // FEBS J. 2013. V. 280. P. 3991–4003.
Van Doren M., Ellis H., Posakony J. The Drosophila extramacrochaetae protein antagonizes sequence-specific DNA binding by daughterless achaete-scute protein complexes // Development. 1991. V. 113. P. 245–255.
Van Doren M., Powell P., Pasternak D. et al. Spatial regulation of proneural gene activity: auto- and cross-activation of achaete is antagonized by extramacrochaetae // Genes Dev. 1992. V. 6. P. 2592–2605.
Venuti J., Goldberg L., Chakraborty T. et al. A myogenic factor from sea urchin embryos capable of programming muscle differentiation in mammalian cells // Proc. Natl. Acad. Sci. U.S.A. 1991. V. 88. P. 6219–6223.
Weintraub H., Dwarki V., Verma I. et al. Muscle-specific transcriptional activation by MyoD // Genes Dev. 1991. V. 5. P. 1377–1386.
White J., Scaffidi A., Davies M. et al. Myotube formation is delayed but not prevented in myoD-deficient skeletal muscle: studies in regenerating whole muscle grafts of adult mice // J. Histochem. Cytochem. 2016. V. 48. № 11. P. 1531–1544.
Zhou Z., Bornemann A. MRF4 protein expression in rege-nerating rat muscle // J. Muscle Res. Cell Motil. 2001. V. 22. P. 311–316.
Дополнительные материалы отсутствуют.