Генетика, 2023, T. 59, № 8, стр. 870-887
Генетические полиморфизмы, ассоциированные с эффективностью коррекции массы тела: систематический обзор
Э. С. Егорова 1, *, И. И. Ахметов 1, 2
1 Казанский государственный медицинский университет
420012 Казань, Россия
2 Научно-исследовательский институт спорта и физических упражнений,
Ливерпульский университет им. Джона Мурса
L3 5AF Ливерпуль, Великобритания
* E-mail: jastspring@yandex.ru
Поступила в редакцию 14.02.2023
После доработки 10.03.2023
Принята к публикации 20.03.2023
- EDN: XSUABK
- DOI: 10.31857/S0016675823080052
Аннотация
Индивидуальные особенности человека не только обусловливают различия в массе тела, но и детерминируют реакцию организма на диету и двигательную активность. Цель данного систематического обзора – описание генетических маркеров, ассоциированных со снижением массы тела в ответ на немедикаментозные методы лечения ожирения, диетотерапию и физические нагрузки. Приемлемые для включения в систематический обзор интервенционные исследования содержали все необходимые параметры генетического полиморфизма, диеты, физической нагрузки и изменений антропометрических или композиционных показателей тела. 91 статья соответствовала критериям и была включена в данный систематический обзор. Подавляющее число исследований (n = 88) было проведено с использованием ген-кандидатного подхода и только три работы выполнены с применением полногеномного поиска ассоциаций (GWAS). Всего было обнаружено 98 генетических вариантов, из которых 72 маркера ассоциированы с эффективностью диетотерапии и 26 – с индивидуальным ответом на физические нагрузки. Следует отметить, что значимость маркеров была подтверждена независимыми исследованиями только для 10 из 98 генетических вариантов. В ближайшие годы следует ожидать прогресса в этом направлении, результатом которого станет разработка метода индивидуального подбора каждому пациенту типа диеты и физической нагрузки для профилактики и лечения ожирения.
Ожирение – одна из актуальных проблем в системе здравоохранения в настоящий момент. Темпы распространенности ожирения во всем мире таковы, что за последние 50 лет оно достигло масштабов пандемии. По данным ВОЗ, количество взрослых в возрасте 18 лет и старше с избыточной массой тела во всем мире составляет около двух миллиардов, из них 650 миллионов страдают ожирением [1, 2]. При этом в России распространенность избыточной массы тела достигает 55%, а ожирения – 26% [3].
Динамика ожирения во многом объясняется современным образом жизни, который характеризуется несбалансированным питанием с высоким содержанием калорий и низким уровнем физической активности, недостаточным для компенсации избыточного потребления. Кроме того, к значимым факторам развития ожирения можно отнести социально-экономический статус, различные поведенческие аспекты [4], микробиом [5], а также измененные в результате технологических достижений циркадные ритмы [6]. Однако, несмотря на общую тенденцию современного образа жизни, люди, живущие в одной и той же среде, демонстрируют значительную вариабельность массы тела, в основе которой помимо средовых лежат и генетические факторы. Так, близнецовые исследования показали, что в предрасположенности человека к ожирению важную роль играют гены, вклад которых оценивается в пределах 40–80% [7, 8]. С учетом последних данных, С. Bouchard пришел к выводу, что генетический компонент индекса массы тела (ИМТ) составляет от 40 до 50%; при этом наследуемость ИМТ и других фенотипов ожирения ниже среди лиц с нормальным весом (30–35%) и выше среди лиц с ожирением (60–80%) [9].
В последнее десятилетие огромный прогресс в идентификации генетических локусов, связанных с ожирением, был достигнут благодаря исследованиям по полногеномному поиску ассоциаций (GWAS – genome wide association study) [10–12]. Недавний крупномасштабный метаанализ GWAS-исследований в европейской популяции выявил 906 локусов, на которые приходится 6% вариабельности ИМТ [13]. При этом размер эффекта каждого аллеля может оцениваться в несколько граммов жировой массы или меньше [14]. Следует отметить, что вклад разных генетических маркеров в риск развития ожирения неодинаков [9]. Предполагается, что за счет увеличения выборок до нескольких миллионов человек в ближайшее время будет обнаружено дополнительно несколько тысяч распространенных генетических вариантов (с частотой >5%), которые будут объяснять до 30% дисперсии ИМТ [14–16]. При этом на оставшуюся долю наследуемости ИМТ, по-видимому, будут приходиться низкочастотные (частота 1–5%) и редкие генетические варианты (≤1%) [17].
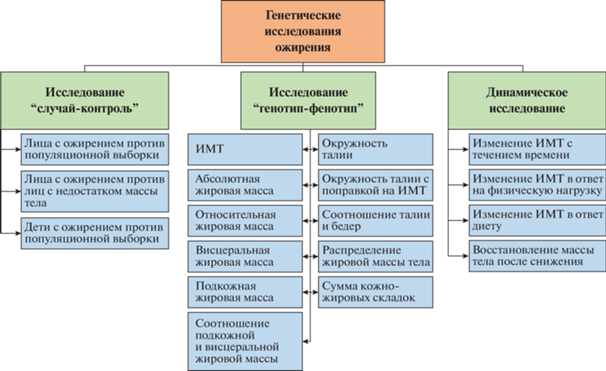
Для обнаружения геномных предикторов ожирения помимо ИМТ исследуются разные фенотипы ожирения, такие как абсолютная и относительная жировая масса, висцеральная и подкожная жировая масса, соотношение висцеральной и подкожной жировой массы, окружность талии, соотношение окружностей талии и бедер и т.п. (рис. 1). Для определения генетической архитектуры ожирения в GWAS-исследованиях используются разные методологические подходы, такие как “случай–контроль”, “генотип–фенотип”, лонгитюдное исследование, где выявляют изменение ИМТ с течением времени. Кроме того, появляется все больше работ, направленных на выявление генетических маркеров эффективности снижения веса в ответ на физические нагрузки, диетотерапию и прием фармакологических препаратов (так называемые интервенционные исследования).
Таким образом, целью данного систематического обзора является описание и анализ доступной на сегодняшний момент информации о генетических маркерах, ассоциированных с эффективностью применения физических нагрузок и диетотерапии для коррекции массы тела.
МАТЕРИАЛЫ И МЕТОДЫ
Стратегия поиска
Поиск литературы и анализ данных проводились в соответствии с рекомендациями PRISMA в электронной базе PubMed среди англоязычной литературы без ограничения по дате публикации по ключевым словам: “полиморфизм”, “SNP”, “генотип”, “диета”, “питание”, “физическая активность”, “физические упражнения”, “тренировка”, “снижение веса”, “снижение жировой массы тела”. Поиск в базе данных PubMed проводили с охватом статей, проиндексированных не позднее 8 декабря 2022 г. Во время поиска фильтры не применялись.
Выбор исследований
Включение статей в данный обзор проводилось в соответствии со следующими критериями: интервенционное исследование, описанная диета или физическая нагрузка, указанная продолжительность диеты или физической нагрузки, наличие данных генетического полиморфизма, наличие результата в виде изменения антропометрических показателей или состава тела. Критериями для исключения являлись неинтервенционные лонгитюдные исследования, исследования, проводившиеся не на людях, а также работы с участием испытуемых, страдающих тяжелым заболеванием (например, раком). Кроме того, были исключены исследования с участием детей, спортсменов, беременных и кормящих женщин, а также пациентов, перенесших хирургические операции.
Извлечение данных
Выявленные исследования из баз данных были извлечены в Microsoft Excel 2016 и автоматически проверены на наличие дубликатов. Оставшиеся после удаления дубликатов статьи были проверены на основе заголовка, аннотации и полного текста в два последовательных этапа. Затем полные статьи оценивались на предмет соответствия требованиям для включения с использованием критериев включения и исключения, изложенных ниже.
Следующие данные были извлечены из полнотекстовых вариантов публикаций: характеристика выборки (количество испытуемых, пол, этническая принадлежность), генетический полиморфизм (название гена и rs-номер), аллель/генотип, ассоциированный с высокой эффективностью вмешательства, характеристика интервенционного вмешательства (тип диетотерапии или физической нагрузки и продолжительность вмешательства), результаты вмешательства (измеряемый параметр, количественное измерение измененяемого параметра у генотипов, уровень значимости P) и ссылка на статью.
Оценка качества исследований (оценка риска предвзятости)
Оценку риска систематической ошибки для рандомизированных контролируемых исследований проводили при помощи адаптированной валидированной версии вопросника Кокрановского сотрудничества [18, 19]. Критерии оценки исследований были следующими: метод рандомизации участников исследования в группы и сокрытие рандомизационной последовательности, “ослепление” исследуемых, медицинского персонала и исследователей, оценивающих эффект вмешательства, пропуски в данных об исходах, неполное предоставление результатов, а также другие источники систематических ошибок (например, конфликт интересов). Риск систематической ошибки для каждого критерия оценивался как низкий, высокий или неясный.
Риск систематической ошибки для публикаций когортных исследований оценивали в соответствии со шкалой оценки качества Ньюкасла–Оттавы [20, 21]. Критерии оценки исследований касались формирования когорт, сопоставимости когорт и оценки исходов, включая восемь подпунктов, которые дают максимальную оценку 9 баллов. В зависимости от итоговых баллов публикации классифицировались: как исследования, у которых высокий риск систематических ошибок (0–5 баллов); исследования, у которых средний риск систематических ошибок (6–7 баллов); исследования, у которых низкий риск систематических ошибок (8–9 баллов).
Кроме того, была проведена оценка методологического качества публикаций в соответствии с критериями, важными для исследований, изучающих генетическую ассоциацию [22, 23]. Оценка качества таких исследований основывалась на восьми критериях: взаимодействие как основная цель исследования, статистический тест на взаимодействие, поправка на множественное тестирование, поправка на этническую принадлежность или стратификацию населения, тестирование равновесия Харди–Вайнберга, тест на групповое сходство на исходном уровне, размер выборки или анализ мощности и достаточное количество указанных деталей процедуры исследования (Приложение) . На основании положительных (+1 балл), нейтральных (0 баллов) или отрицательных (–1 балл) оценок по каждому пункту общее количество баллов за каждую публикацию может варьироваться от –8 до 8 баллов. Таким образом, статьи, набравшие от 6 до 8 баллов, оценивались как исследования с высоким методологическим качеством; статьи, набравшие от 2 до 5 баллов, оценивались как исследования со средним качеством, а статьи с –8 до 1 балла – как исследования с низким качеством.
РЕЗУЛЬТАТЫ
Отбор и характеристика исследований
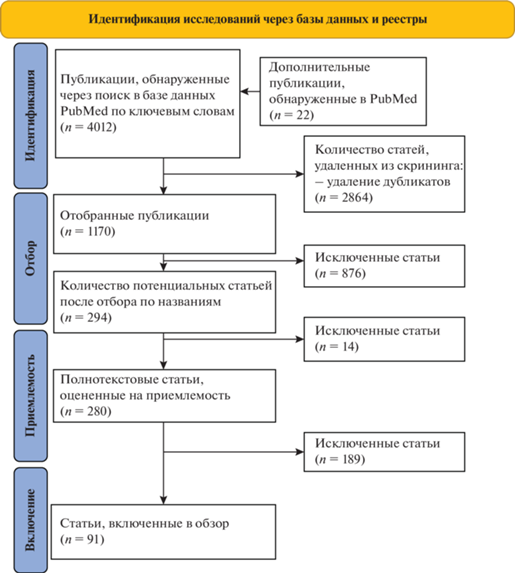
Первоначальный поиск, по ключевым словам, в базе данных PubMed выявил 4012 публикаций (рис. 2). Кроме того, были дополнительно включены в анализ 22 публикации, обнаруженные по поиску похожих статей и не найденные в результате поиска по ключевым словам. После удаления дубликатов (n = 2864) 1170 уникальных статей были отобраны для анализа в соответствии с темой систематического обзора по названию и аннотации публикации, из которых 876 статей были исключены. Затем для подробной оценки было прочитано 280 полнотекстовых вариантов исследований, из которых 189 статей были исключены по причине: исходов, не соответствующих коррекции массы тела (n = 106); статистически незначимых различий в изменении антропометрических данных или данных состава тела между генотипами (n = 34); неинтервенционных исследований (n = 17); исследований, изучавших влияние негенетических полиморфизмов на коррекцию массы тела (n = 13); неуказанных параметров вмешательства (n = 5); медикаментозных или хирургических вмешательств (n = 4); исследований, в которых не было единых параметров вмешательств для всех испытуемых (n = 3), испытуемых спортсменов (n = 3), детей и подростков (n = 2); неуказанного уровня значимости P (n = 1); отсутствия полнотекстового варианта публикации (n = 1).
В общей сложности 91 интервенционное исследование соответствовало нашим критериям и было включено в данный систематический обзор. Период публикации включенных статей колебался с 1997 по 2022 г. Общее количество генетических полиморфизмов из выбранных статей составило 98 (из них уникальных 88). Во всех вмешательствах и генетических скринингах участвовало 23 135 человек, при этом размер выборки варьировал от 17 до 1995 участников. Продолжительность вмешательств варьировала от двух недель до трех лет. Исследования различались по характеристикам участников, а также по типу диетического вмешательства, типу физической нагрузки (ФН) и по продолжительности вмешательства (Приложение) .
Качество исследования и риск систематической ошибки
Кокрановская шкала оценки риска систематической ошибки показала низкий риск для 14 рандомизированных контролируемых исследований, средний риск для шести исследований и высокий риск для десяти исследований (Приложение) . Причиной низкой оценки рандомизированных контролируемых исследований является характер вмешательства в образ жизни, усложняющего “ослепление” пациента и персонала.
Оценка систематических ошибок нерандомизированных контролируемых исследований по шкале оценки качества Ньюкасла–Оттавы выявила 48 исследований с низким риском и 13 исследований со средним риском (Приложение) . Основными причинами снижения баллов было отсутствие информации о дополнительных критериях сопоставимости когорт, а именно статистически значимых различий по приверженности диете и уровню физических нагрузок между исследуемыми.
Оценка методологического качества публикаций в соответствии со специально разработанной шкалой для исследований, изучающих ассоциацию генов с диетой/физической нагрузкой, продемонстрировала 56 публикаций с высоким методологическим качеством и 35 публикаций со средним методологическим качеством (Приложение) . Основными причинами снижения качества публикаций стало отсутствие поправки на множественное сравнение, отсутствие поправки на стратификацию населения и недостаточный размер исследуемой выборки.
Подавляющее число исследований (n = 88) было проведено с использованием ген-кандидатного подхода и только три работы выполнены с применением полногеномного поиска ассоциаций (GWAS). Всего было обнаружено 98 генетических вариантов, из которых 72 маркера ассоциированы с эффективностью диетотерапии и 26 – с индивидуальным ответом на физические нагрузки. Следует отметить, что значимость маркеров была подтверждена независимыми исследованиями только для 10 из 98 генетических вариантов.
Генетические маркеры эффективности диетотерапии в лечении ожирения
В 67 исследованиях было обнаружено 72 уникальных генетических маркера: 27 генетических вариантов ассоциировались с эффективностью коррекции массы тела (МТ) в ответ на умеренно гипокалорийную диетотерапию, 10 генетических маркеров – в ответ на выраженно гипокалорийную диету, 18 – в ответ на гипокалорийную низкожировую диету, 18 – в ответ на гипокалорийную высокобелковую диету, 6 – в ответ на гипокалорийную низкоуглеводную диету, 8 – в ответ на средиземноморскую диету, 4 – в ответ на гипокалорийную диету, обогащенную мононенасыщенными и/или полиненасыщенными жирами, и 1 генетический маркер – в ответ на гипокалорийную диету, обогащенную клетчаткой.
Наиболее значимые данные были получены в результате проведенных GWAS-исследований с участием большого числа индивидов с избыточной массой тела и ожирением. Необходимо отметить исследование М. Nikpay с соавт., в котором носители аллеля дикого типа С полиморфизма rs679482 гена SGCG (Sarcoglycan gamma – саркогликан гамма) были более успешными в снижении МТ в ответ на гипокалорийную высокобелковую диету в течение 12 нед. [24] (Приложение) . Еще одно крупное GWAS-исследование выявило четыре генетических маркера (полиморфизм rs6981587 гена ANK1/MIR486-2 (Аnkyrin-1/MicroRNA 486-2 – анкирин 1/микроРНК 486-2), полиморфизмы rs873822, rs870879, rs1027493 гена RBSG4 (Long intergenic non-protein coding RNA 1363 – длинная межгенная небелок-кодирующая РНК 1363)), которые ассоциировались с эффективностью снижения МТ в ответ на гипокалорийную высокобелковую диету в течение 16 нед. [25].
В интервенционных многоцентровых исследованиях с участием свыше 1000 испытуемых было выявлено, что средиземноморская диета эффективна для коррекции МТ для носителей рискового аллеля G полиморфизма rs1801282 гена PPARG (Peroxisome proliferator – activated receptor gamma – гамма-рецептор, активируемый пролифератором пероксисом) [26], носителей минорного аллеля С полиморфизма rs2289487 гена PLIN1 (Perilipin 1 – перилипин 1) [27], носителей генотипа ТТ полиморфизма rs1052700 гена PLIN1 [27] и носителей генотипа АА полиморфизма rs1801260 гена CLOCK (Circadian locomoter output cycles protein kaput – регулятор циркадных ритмов) [28]. В исследовании J. Aberle с соавт. [29] была продемонстрирована эффективность низкожировой диеты в течение 12 нед. для носителей минорного аллеля А полиморфизма rs1049353 гена CB1 (Cannabinoid receptor 1 – каннабиноидный рецептор типа 1) и носителей протективного аллеля G полиморфизма rs1862513 гена RETN (Resistin – резистин) [29].
При этом значимое влияние на коррекцию МТ как минимум в двух исследованиях показали 13 полиморфизмов генов: rs2419621 гена ACSL5 (Acyl-coA synthetase long chain family member 5 – член семейства длинноцепочечных ацил-КоА-синтетаз 5), rs10182181 гена ADCY3 (Adenylate сyclase 3 – аденилатциклаза типа 3), rs4994 гена ADRB3 (Adrenoceptor beta 3 – бета-3-адренергический рецептор), rs1801260 гена CLOCK, rs1799883 гена FABP2 (Fatty acid binding protein 2 – белок, связывающий жирные кислоты 2), rs9939609 гена FTO (Fat mass and obesity-associated protein – альфа-кетоглутарат-зависимая диоксигеназа), rs2943641 гена IRS1 (Insulin receptor substrate 1 – cубстрат инсулинового рецептора 1), rs2605100 гена LYPLAL1 (Lysophospholipase like 1 – лизофосфолипазоподобный белок 1), rs10830963 гена MTNR1B (Melatonin receptor 1B – Мелатониновый рецептор типа 1B), rs894160 гена PLIN1, rs1801282 гена PPARG, rs7903146 гена TCF7L2 (Transcription factor 7 Like 2 – Транскрипционный фактор 7-подобный 2) и rs1800849 гена UCP3 (Uncoupling protein 3 – митохондриальный разобщающий белок 3). Важно отметить, что восемь из 13 полиморфизмов генов продемонстрировали значимое влияние на коррекцию МТ в ответ на один и тот же тип диетотерапии: rs10182181 гена ADCY3, rs4994 гена ADRB3, rs1801260 гена CLOCK, rs2943641 гена IRS1, rs2605100 гена LYPLAL1, rs10830963 гена MTNR1B, rs894160 гена PLIN1, rs1801282 гена PPARG.
Так, в двух исследованиях было показано, что носители рискового аллеля G полиморфизма rs10182181 гена ADCY3 значимо больше снижали жировую массу тела (ЖМТ) [30] и ИМТ [31] в ответ на гипокалорийную низкожировую диету. М. Garaulet с соавт. [28] в интервенционном исследовании с большим числом испытуемых (n = 1495) подтвердили, что средиземноморская диета является наиболее эффективной для носителей рискового аллеля А полиморфизма rs1801260 гена CLOCK [32]. Также репликационными исследованиями было подтверждено, что для носителей протективного аллеля А полиморфизма rs4994 гена ADRB3 [33, 34] и носителей аллеля G полиморфизма rs894160 гена PLIN1 [35, 36] эффективной при коррекции массы тела является гипокалорийная диета. Для носителей протективного аллеля С полиморфизма rs2943641 гена IRS1 эффективной является низкожировая диета [31, 37], для носителей протективного аллеля G полиморфизма rs2605100 гена LYPLAL1 [31, 38] и носителей аллеля С полиморфизма rs10830963 гена MTNR1B [39, 40] – высокобелковая диета, для носителей рискового аллеля G полиморфизма rs1801282 гена PPARG – средиземноморская диета [26, 41].
По данным ряда исследований было выявлено, что носители одних и тех же аллелей могут одинаково эффективно снижать МТ (или другие антропометрические показатели, или показатели состава тела) при разных типах диетотерапий. Как гипокалорийная диета в течение 24 нед. [42], так и менее продолжительная гипокалорийная высокобелковая диета в течение 12 нед. [43] эффективно влияет на снижение МТ и состав тела для носителей альтернативного аллеля Т полиморфизма rs2419621 гена ACSL5. Такое же явление характерно и для полиморфизмов гена CB2R (Cannabinoid receptor 2 – каннабиноидный рецептор 2) [44], гена FABP2 [45, 46], гена FTO [47–50], гена MC4R (Melanocortin 4 receptor – рецептор меланокортина 4) [51, 52], гена MTNR1B [39, 53–55], гена PPARG [26, 41], гена TCF7L2 [38, 56], гена UCP2 (Uncoupling protein 2 – митохондриальный разобщающий белок 2) [31, 57].
В результате исследований было обнаружено, что полиморфизм rs987237 гена TFAP2B (Transcription factor AP-2 beta – транскрипционный фактор АР-2 бета) модифицировал влияние диеты на коррекцию веса в зависимости от состава макронутриентов: для носителей рискового аллеля А предпочтительной для эффективного снижения МТ является гипокалорийная низкоуглеводная диета, но не низкожировая [58]. Аналогичная картина наблюдалась для полиморфизма rs7957197 гена HNF1A (Hepatocyte nuclear factor 1-alpha – ядерный фактор гепатоцитов 1-альфа), где носители альтернативного аллеля А значимо больше снижали МТ при гипокалорийной низкожировой диете, а носители аллеля Т – напротив, при низкоуглеводной [59]. Носители генотипа СС полиморфизма rs1801133 гена MTFHR (Methylenetetrahydrofolate reductase – метилентетрагидрофолатредуктаза) эффективно снижали МТ в ответ на средиземноморскую диету [60], а носители генотипа АА – в ответ на высокобелковую [31].
Генетические маркеры эффективности физических нагрузок при лечении ожирения
В 24 исследованиях обнаружены 26 генетических маркеров, ассоциированных со снижением МТ в ответ на различные типы ФН. Из них 22 ассоциировались с эффективностью снижения МТ в ответ на аэробную тренировку, 2 – в ответ на силовую, 1 – в ответ на интервальную и 2 маркера – в ответ на ФН без уточнения типа.
Наиболее значимые результаты были продемонстрированы в GWAS-исследовании с участием 126 польских женщин, вовлеченных в аэробную 12-недельную тренировку [61], согласно которому носители редкого Т аллеля (частота в европейской популяции около 1.5%) полиморфизма rs116143768 гена ACSL1 (Acyl-CoA synthetase long chain family member 1 – длинноцепочечная ацил-КоА-синтетаза 1) значимо больше снижали относительную ЖМТ (P = 1.18 × 10–9) (Приложение) . Одни из наиболее значимых результатов по эффективности снижения ЖМТ в ответ на аэробные нагрузки высокой интенсивности были получены I. Mazur с соавт. [62] для носителей аллеля Т полиморфизма rs1765040 гена PPARGC1A (Peroxisome proliferator-activated receptor gamma coactivator 1-alpha – гамма-коактиватор рецептора, активируемого пролифератором пероксисом 1-альфа) (P = 0.00013).
Из описанных в данном систематическом обзоре 26 генетических маркеров значимое влияние на коррекцию МТ в ответ на ФН как минимум в двух независимых исследованиях продемонстрировали только двa генетических полиморфизмa: rs8050136 гена FTO и rs1801282 гена PPARG. Так репликационными исследованиями было показано, что носители рискового аллеля A полиморфизма rs8050136 гена FTO значимо больше снижали МТ в ответ на аэробные нагрузки в течение шести месяцев у женщин (n = 234) [63], а также в течение 12 нед. только у мужчин (n = 101) [64]. В исследовании Т. Østergård с соавт. [65] носители аллеля G полиморфизма rs1801282 гена PPARG продемонстрировали эффективное снижение МТ в ответ на аэробные нагрузки в течение десяти недель. В дальнейшем эти результаты были подтверждены исследованием Р. Franks с соавт. [66] с участием 1004 испытуемых с избыточной массой тела или ожирением, в котором носители аллеля G значимо больше снижали МТ и ЖМТ в ответ на преимущественно аэробную ФН умеренной интенсивности в течение одного года.
Силовые тренировки с отягощением в течение 12 нед. продемонстрировали свою эффективность при коррекции МТ для носителей генотипа GG полиморфизма rs7566605 гена INSIG2 (Insulin induced gene 2 – инсулин-индуцированный белок 2) в исследовании с участием 752 испытуемых [67]. J. Cameron с соавт. [68] в работе с участием 127 женщин с избыточной массой тела или ожирением показали, что носители протективного аллеля С полиморфизма rs1800497 гена DRD2 (Dopamine receptor D2 – дофаминовый D2-рецептор) более успешны в снижении массы тела, жировой массы тела и ИМТ при более продолжительных силовых нагрузках в течение шести месяцев. В статье O. Andrade-Mayorga c cоавт. [69] было выявлено, что высокоинтенсивные интервальные тренировки в течение 12 нед. эффективны в снижении ЖМТ для носителей рискового аллеля Т полиморфизма rs1052700 гена PLIN1.
Наряду с этим были обнаружены противоречивые результаты. Так, в исследовании D. Phares с соавт. [70] с участием 70 человек носители аллеля G полиморфизма rs4994 гена ADRB3 эффективно снижали ЖМТ, а D. de Luis с соавт. [71], напротив, продемонстрировали наибольшую эффективность для носителей генотипа АА.
ОБСУЖДЕНИЕ
В данном систематическом обзоре было рассмотрено 91 исследование, выявляющее ассоциацию генетических вариантов с эффективностью коррекции массы тела: в 67 работах изучалось влияние диеты на коррекцию массы тела и в 24 статьях – влияние физической нагрузки. Несмотря на то что нутриенты обладают существенным модулирующим эффектом на ожирение, физическая нагрузка также может привести к значительной коррекции массы тела. При этом при физической активности не только снижается жировая масса тела, а также может увеличиться мышечная масса, рост которой также способствует улучшению обмена веществ и повышению липолиза [72]. Необходимо увеличение числа экспериментальных работ по изучению влияния разных типов физической активности на коррекцию массы тела в зависимости от генотипа человека. Данный систематический обзор представляет результаты 67 исследований, изучавших ассоциацию 72 генетических маркеров с коррекцией массы тела в ответ на диетотерапию. В большинстве публикаций (n = 27) исследовалась эффективность сбалансированной гипокалорийной диеты, что скорее всего связано с тем, что это наиболее простой и эффективный способ коррекции массы тела. Однако было показано, что макронутриентный состав пищевого рациона влияет на гормональный фон, метаболические пути, экспрессию генов, а также на состав микробиома кишечника [73]. Поэтому необходимо отметить актуальность проведения исследований, изучающих эффективность различных по составу макронутриентов диет на коррекцию массы тела в зависимости от генетического статуса индивида.
Анализ результатов выявил, что только восемь генетических маркеров подтвердили свою ассоциацию с эффективностью одной и той же диетотерапии в репликационных исследованиях: rs10182181 гена ADCY3, rs4994 гена ADRB3, rs1801260 гена CLOCK, rs2943641 гена IRS1, rs2605100 гена LYPLAL1, rs10830963 гена MTNR1B, rs894160 гена PLIN1 и rs1801282 гена PPARG.
В двух исследованиях было продемонстрировано, что носители рискового аллеля G полиморфизма rs10182181 гена ADCY3 эффективнее снижали ЖМТ или ИМТ в ответ на низкожировую диету, но не на высокобелковую диету [30, 31]. Известно, что ген ADCY3 кодирует фермент аденилатциклазу типа 3, который превращает АТФ в цАМФ. Данный фермент (ADCY3) участвует в большом количестве физиологических метаболических процессов, включая регуляцию углеводного и липидного обменов, а также развитие и функцию жировой ткани, а также регулирует экспрессию генов, участвующих в адипогенезе, термогенезе и липолизе [74]. На основании данных, полученных в экспериментах на мышах, известно, что гаплонедостаточность гетерозиготных мышей по гену Adcy3 +/– приводит к снижению экспрессии генов, участвующих в термогенезе, окислении жирных кислот и передаче сигналов инсулина у мышей, и, наоборот, к усилению экспрессии генов, связанных с адипогенезом, в периферических тканях [75]. С другой стороны, диета с высоким содержанием жиров приводит к снижению экспрессии гена аденилатциклазы типа 3 [76]. Кроме того, по данным GTEx Portal [77] аллель G ассоциируется с более высоким уровнем экспрессии ADCY3 в крови и жировой ткани. На основании этих данных можно предположить, что низкожировая диета вероятно будет способствовать повышению экспрессии гена ADCY3, снижению адипогенеза и улучшению регулирования уровня инсулина в организме. Однако механизмы, лежащие в основе модулирования потребления макронутриентов генетическим вариантом ADCY3, до конца не изучены, и необходимы дальнейшие экспериментальные исследования.
Носители протективного аллеля С полиморфизма rs2943641 гена IRS1 наиболее успешно снижают МТ при низкожировой диете, но не при низкоуглеводной или высокобелковой [31, 37]. Потенциальные механизмы, лежащие в основе этих результатов, неизвестны, но могут быть связаны с резистентностью к инсулину, индуцированной липидами [78].
Носители протективного аллеля G полиморфизма rs2605100 гена LYPLAL1 и носители аллеля С полиморфизма rs10830963 гена MTNR1B эффективно снижали МТ в ответ на высокобелковую диету. Интересно, что в исследованиях О. Ramos-Lopez с соавт. [31, 38] носители протективного аллеля G полиморфизма rs2605100 гена LYPLAL1 эффективно снижали жировую массу тела и ИМТ в ответ на гипокалорийную высокобелковую, но не в ответ на гипокалорийную низкожировую. При этом известно, что ген LYPLAL1 кодирует лизофосфолипазоподобный белок 1, действующий как триглицеридлипаза, а аллель G полиморфизма гена LYPLAL1, по всей видимости, связан с повышенной экспрессией гены липазы и повышенными концентрациями триглицеридов в сыворотке натощак, что приводит к развитию ожирения [79]. К настоящему моменту биохимическая роль LYPLAL1 до конца не установлена [80], поэтому необходимы дополнительные исследования, которые бы выявили механизм действия лизофосфолипазоподобного белка.
В нескольких исследованиях было выявлено, что носители протективного аллеля С полиморфизма rs10830963 гена MTNR1B могут эффективно снижать МТ в ответ на разные по составу макронутриентов гипокалорийные диеты [39, 40, 53, 54], а носители рискового аллеля G – в ответ на гипокалорийную низкожировую [55]. Установлено, что ген MTNR1B кодирует рецептор мелатонина, экспрессируемый в супрахиазматическом ядре, центре контроля циркадных ритмов, а также в β-клетках поджелудочной железы [81, 82]. Было обнаружено, что диета с высоким содержанием жиров модифицирует циркадные ритмы человека [83, 84], что ведет к значительному изменению циркадной ритмичности различных гормонов, связанных с ожирением. При этом полиморфизм rs10830963 гена MTNR1B может участвовать в регуляции экспрессии гена MTNR1B или экспрессии других генов, которые могут влиять на роль мелатонина в энергетическом балансе.
В двух исследованиях была продемонстрирована эффективность средиземноморской диеты для носителей рискового аллеля A полиморфизма гена CLOCK [28, 32] и рискового аллеля G полиморфизма rs1801282 гена PPARG [26, 41]. Кодируемый геном CLOCK циркадный осциллятор играет важную роль в развитии ожирения и метаболического синдрома [85]. Выявлено, что носители аллеля G могут проявлять большую степень ожирения и испытывать большие трудности с уменьшением веса в ответ на низкокалорийную диету. Это может быть связано с особенностями циркадных ритмов и пищевым поведением человека, что вероятно затрудняет коррекцию массы тела. Тем не менее было показано, что более длительное соблюдение средиземноморской диеты связано с улучшением качества сна [86] и большей эффективностью коррекции МТ [87, 88].
В исследовании A. Chmurzynska c соавт. [41] носители рискового аллеля G полиморфизма rs1801282 гена PPARG одинаково успешно снижали МТ в ответ на гипокалорийную сбалансированную и гипокалорийную средиземноморскую диеты. Однако коррекция массы тела в ответ на гипокалорийную диету у носителей аллеля G сопровождалась значительным снижением безжировой массы тела. Напротив, в ответ на гипокалорийную средиземноморскую диету наблюдалось значимое снижение абдоминальной жировой массы тела у индивидов с таким же генотипом. Данное наблюдение подтвердило результаты М. Garaulet с соавт. [26]. Известно, что ненасыщенные жирные кислоты, которыми богата средиземноморская диета, являются лигандами для гамма-рецепторов, активируемых пролифератором пероксисом [89]. Активация PPARG жирными кислотами опосредует экспрессию нескольких генов-мишеней, участвующих в накоплении жировой ткани, таких как липопротеинлипаза, а также играет роль в насыщении жировой триглицеридлипазы [89]. Кроме того, сообщалось о защите от ожирения, вызванного пищевым жиром, и резистентности к инсулину у мышей с дефицитом Pparg (Pparg +/–) [90].
Также было выявлено, что полиморфизм rs894160 гена PLIN1 ассоциировался с изменением массы тела в ответ на краткосрочную (в течение 12 нед.) и длительную (в течение одного года) гипокалорийную диету [35, 36]. Перилипин участвует в катехоламинстимулированном липолизе посредством взаимодействия с липазой [91]. Было продемонстрировано, что носители аллеля дикого типа G более успешны в снижении массы тела в ответ на ограничение калорийности в связи с неизменными уровнями перлипина и скорости окисления липидов. Соответственно генетические маркеры обусловливают разную степень окисления жиров в ответ на одинаковую степень отрицательного энергетического баланса и изменения массы тела у индивидов с разными генотипами.
Нами было выявлено 24 генетических полиморфизма, которые показали значимую ассоциацию с коррекцией веса в ответ на физические нагрузки. При этом репликационные исследования, подтвердившую данную ассоциацию, были проведены только для двух полиморфизмов генов: FTO [63, 64] и PPARG [65, 66].
J. Mitchell с соавт. [63] продемонстрировали, что полиморфизм rs8050136 гена FTO ассоциируется со снижением МТ в ответ на аэробную нагрузку умеренной интенсивности в течение шести месяцев. При этом значимо больше снижали массу тела носители рискового аллеля А данного варианта. Эти данные были подтверждены в исследовании W. Wang с соавт. [64] на китайской популяции, где также носительство аллеля А среди мужчин благоприятно влияло на снижение массы тела. Однако в исследовании HERITAGE с участием 481 человека была показана обратная картина: носители протективного аллеля С значимо больше по сравнению с носителями рискового аллеля А снижали жировую массу тела в ответ на аэробную нагрузку низкой/умеренной интенсивности [92].
Известно, что физическая нагрузка высокой интенсивности вызывает снижение экспрессии гена FTO в скелетных мышцах по сравнению с физической нагрузкой низкой интенсивности [93]. При этом было выявлено, что у носителей генотипа АА (А – аллель риска) полиморфизма rs9939609 гена FTO уровень глюкозы в мышцах выше в ответ на тренировку. Это может свидетельствовать о метаболическом сдвиге в сторону большего окисления липидов и отхода от окисления глюкозы потенциально за счет активации АМФ-активируемой протеинкиназы и FTO-зависимого деметилирования N6-метиладенозина [94]. Можно предположить, что благодаря этому механизму носители аллеля риска могут получить преимущество в снижении веса в ответ на физическую нагрузку высокой или умеренной, но не низкой интенсивности.
Кроме того, в обсервационных продолжительных исследованиях было продемонстрировано, что рисковый аллель А полиморфизма rs9939609 гена FTO в меньшей степени повышал риск развития ожирения (на 30%) в группе физически активных людей, чем в группе малоподвижных людей [95–97], а по данным некоторых исследований – на 80% [98, 99].
Исследование Т. Østergård с соавт. [65] об ассоциации рискового аллеля G полиморфизма rs1801282 гена PPARG со значимо большим снижением массы тела в ответ на физическую нагрузку подтвердилось многоцентровым исследованием с участием свыше 1000 человек [66]. Однако в публикации А. Zarebska с соавт. [100] сообщили о том, что носители протективного аллеля С полиморфизма гена PPARG значимо больше снижают жировую массу тела по сравнению с носителями аллеля G. Как упоминалось выше, PPARG регулирует адипогенез, липолиз и чувствительность к инсулину. При этом в недавнем метаанализе с участием 70 317 человек сообщается, что аллель G полиморфизма rs1801282 гена PPARG обусловливает повышенный риск ожирения [101]. Однако в некоторых исследованиях было выявлено, что аллель G ассоциируется с высоким ИМТ только у лиц с выраженным ожирением, а у индивидов с нормальной массой тела эта взаимосвязь ослабевает [102].
Возможно этим объясняются противоречивые результаты, поскольку известно, что в исследовании А. Zarebska с соавт. [100] средний ИМТ испытуемых составлял 21.5 ± 2.5 кг/м2, а в исследованиях Т. Østergård с соавт. [65] и Р. Franks с соавт. [66] средний ИМТ был выше, 25.7 ± 2.7 и 34.1 кг/м2 соответственно. Кроме того, возможно более молодые участники исследования А. Zarebska с соавт. [100] имели исходно более высокий уровень физической активности, чем испытуемые в исследованиях Т. Østergård с соавт. [65] и Р. Franks с соавт. [66], который мог модифицировать ассоциацию аллеля G c эффективностью коррекции веса.
Недостаточное количество воспроизведенных данных в репликационных исследованиях вероятно связано с отсутствием поправки на множественное сравнение в некоторых публикациях, которое могло привести к ложноположительным результатам. Однако необходимо сказать, что поправка на множественное сравнение в выборке размер которой часто бывает ограничен в силу характера проводимых вмешательств, также может привести к возникновению ошибки второго рода, т.е. к невыявлению истинной взаимосвязи. Кроме того, обнаруженные противоречивые результаты могут быть связаны не только с разными размерами выборок, но и с различием в продолжительности исследований, типах диетического вмешательства или физических нагрузок, с дизайном исследования, а также с этнической принадлежностью участников исследования.
Тем не менее во множестве других исследованиях было продемонстрировано, что питание и двигательная активность оказывают значительное влияние на коррекцию массы тела в зависимости от генотипа индивида. Так, было показано, что влияние потребления свободных сахаров и сахаросодержащих напитков, а также жареной пищи на ИМТ в значительной степени зависит от полигенного профиля, состоящего из 32 полиморфизмов генов, ассоциированных с риском развития ожирения [103, 104]. Ассоциация генетических маркеров с ИМТ была значимее среди участников с более высоким потреблением сахаросодержащих напитков, чем среди участников с более низким потреблением: увеличение ИМТ на каждые десять аллелей риска составило 1.00 кг/м2 при приеме менее одной порции в месяц, 1.20 кг/м2 при приеме от одной до четырех порций в месяц, 1.37 кг/м2 при приеме от двух до шести порций в неделю [103]. В другой работе было показано, что генетическая связь с ожирением усиливалась при более высоком потреблении жареной пищи: риск развития ожирения на каждые 10 аллелей риска увеличивался в 1.61 раза при потребления жареной пищи менее одного раза в неделю, в 2.12 раза при потреблении от одного до трех раз и в 2.72 раза при потреблении более трех раз в неделю. При этом вариант гена FTO показал самое сильное взаимодействие с потреблением жареной пищи [104]. Эти данные свидетельствуют о том, что лица с высоким генетическим риском ожирения оказались более восприимчивыми к негативному влиянию сахаросодержащих напитков и жареной пищи, что привело у них к более выраженному повышению ИМТ.
Модифицирующее влияние на генетическую предрасположенность к ожирению оказывает не только диетический состав макронутриентов, но и частота приемов пищи. Регулярная частота приема пищи ослабляет генетическую предрасположенность к увеличению ИМТ, как с точки зрения одного из исследуемых генетических локусов (rs1421085 гена FTO, rs17782313 гена MC4R, rs6265 гена BDNF, rs10938397 гена GNPDA2, rs1424233 гена MAF, rs6548238 гена TMEM18, rs11084753 гена KCTD15, rs2815752 гена NEGR1), так и на основании полигенного анализа [105]. Кроме того, время приема пищи может играть решающую роль в ожирении, поскольку циркадные ритмы имеют важное значение в энергетическом обмене. Диетический подход, в основе которого лежит ограничение времени приема пищи, представляет собой многообещающий и эффективный метод лечения ожирения без снижения общей калорийности рациона [106, 107].
Большинство рисковых аллелей (ранее показавших связь с риском ожирения), описанных в настоящем обзоре, ассоциируются со сниженной эффективностью коррекции веса. Таким образом, рисковые аллели не только приводят к увеличению жировой массы в ответ на пониженную физическую активность и переедание, но и затрудняют процесс снижения веса в ответ на диету и физические нагрузки.
Подводя итог, следует выделить, что преимуществом данной работы является всесторонний обзор исследований, изучающих генетическую ассоциацию не только с эффективностью диетотерапий, но и эффективностью физических нагрузок по отношению к изменениям массы тела. Кроме того, мы провели оценку систематических ошибок и методологического качества исследований, включенных в данную статью.
В настоящей работе мы рассмотрели 91 исследование, в которых была показана ассоциация 98 генетических маркеров с эффективностью снижения массы тела под действием определенного типа тренировочного режима или диеты. Стоит отметить, что большая часть генетических вариантов, выявленных в результате интервенционных исследований, обнаружена при помощи ген-кандидатного подхода, ограниченного существующими знаниями исследователей о биологии ожирения. Это свидетельствует о том, что на сегодняшний момент наши знания относительно генетических маркеров, влияющих на массу тела и функциональный ответ на физические нагрузки и прием макронутриентов, ограничены. Требуется проведение дальнейших крупномасштабных GWAS-исследований, репликативных исследований и метаанализов, которые позволят выявить новые генетические маркеры, ассоциированные с адаптационными реакциями организма на тренировку и прием макронутриентов.
Выявление новых генетических маркеров позволит объяснить большую фенотипическую дисперсию изменения веса и, в свою очередь, повысит их прогностическую эффективность. Таким образом, настоящий обзор позволил выявить 98 генетических маркеров эффективности снижения массы тела в ответ на разные типы диет и физических нагрузок. В ближайшие годы следует ожидать прогресса в этом направлении, результатом которого станет разработка метода индивидуального подбора каждому пациенту типа диеты и физической нагрузки для профилактики и лечения ожирения.
Публикация подготовлена в рамках научно-исследовательского проекта, поддержанного грантом ФГБОУ ВО Казанский ГМУ Минздрава России на проведение научных исследований в рамках Программы развития Университета (“Разработка диагностического комплекса, направленного на профилактику и лечение ожирения с учетом полиморфизмов генов, ассоциированных с циркадными ритмами человека” № НИР 94-017-2022).
Настоящая статья не содержит каких-либо исследований с использованием в качестве объекта животных.
Настоящая статья не содержит каких-либо исследований с участием в качестве объекта людей.
Авторы заявляют, что у них нет конфликта интересов.
Список литературы
Yanovski J.A. Obesity: Trends in underweight and obesity – scale of the problem // Nat. Rev. Endocrinol. 2018. V. 14. № 1. P. 5–6. https://doi.org/10.1038/nrendo.2017.157
GBD 2017 Diet Collaborators. Health effects of dietary risks in 195 countries, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017 // Lancet. 2019. V. 393. e10184. P. 1958–1972. https://doi.org/10.1016/S0140-6736(19)30041-8
Мартинчик А.Н., Лайкам К.Э., Козырева Н.А. и др. Распространение ожирения в различных социально-демографических группах населения России // Вопр. питания. 2021. Т. 90. № 3(535). С. 67–76.
Narciso J., Silva A.J., Rodrigues V. et al. Behavioral, contextual and biological factors associated with obesity during adolescence: A systematic review // PLoS One. 2019. V. 14. № 4. https://doi.org/10.1371/journal.pone.0214941
Gao R., Zhu C., Li H. et al. Dysbiosis signatures of gut microbiota along the sequence from healthy, young patients to those with overweight and obesity // Obesity (Silver Spring). 2018. V. 26. № 2. P. 351–361. https://doi.org/10.1002/oby.22088
Romo-Nava F., Guerdjikova A.I., Mori N.N. et al. A matter of time: A systematic scoping review on a potential role of the circadian system in binge eating behavior // Front. Nutr. 2022. V. 9. https://doi.org/10.3389/fnut.2022.978412
Silventoinen K., Jelenkovic A., Sund R. et al. Genetic and environmental effects on body mass index from infancy to the onset of adulthood: An individual-based pooled analysis of 45 twin cohorts participating in the COllaborative project of Development of Anthropometrical measures in Twins (CODATwins) study // Am. J. Clin. Nutr. 2016. V. 104. № 2. P. 371–379. https://doi.org/10.3945/ajcn.116.130252
Silventoinen K., Jelenkovic A., Sund R. et al. Genetic and environmental variation in educational attainment: An individual-based analysis of 28 twin cohorts // Sci. Rep. 2020. V. 10. № 1. P. 12681. https://doi.org/10.1038/s41598-020-69526-6
Bouchard C. Genetics of obesity: What we have learned over decades of research // Obesity (Silver Spring). 2021. V. 29. № 5. P. 802–820. https://doi.org/10.1002/oby.23116
Fox C.S., Liu Y., White C.C. et al. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women // PLoS Genet. 2012. V. 8. № 5. e1002695. https://doi.org/10.1371/journal.pgen.1002695
Yengo L., Sidorenko J., Kemper K.E. et al. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry // Hum. Mol. Genet. 2018. V. 27. № 20. P. 3641–3649. https://doi.org/10.1093/hmg/ddy271
Warner E.T., Jiang L., Adjei D.N. et al. A genome-wide association study of childhood body fatness // Obesity (Silver Spring). 2021. V. 29. № 2. P. 446–453. https://doi.org/10.1002/oby.23070
Huang J., Huffman J.E., Huang Y. et al. Genomics and phenomics of body mass index reveals a complex disease network // Nat. Commun. 2022. V. 29. V. 13. № 1. P. 7973. https://doi.org/10.1038/s41467-022-35553-2
Locke A.E., Kahali B., Berndt S.I. et al. Genetic studies of body mass index yield new insights for obesity biology // Nature. 2015. V. 518. № 7538. P. 197–206. https://doi.org/10.1038/nature14177
Ge T., Chen C.Y., Neale B.M. et al. Phenome-wide heritability analysis of the UK Biobank // PLoS Genet. 2018. V. 14. № 2. e1007228. https://doi.org/10.1371/journal.pgen.1006711
Khera A.V., Chaffin M., Wade K.H. et al. Polygenic prediction of weight and obesity trajectories from birth to adulthood // Cell. 2019. V. 177. № 3. P. 587–596. e9. https://doi.org/10.1016/j.cell.2019.03.028
Wainschtein P., Jain D., Zheng Z. et al. Assessing the contribution of rare variants to complex trait heritability from whole-genome sequence data // Nat. Genet. 2022. V. 54. № 3. P. 263–273. https://doi.org/10.1038/s41588-021-00997-7
Higgins J.P., Altman D.G., Gøtzsche P.C. et al. The Cochrane Collaboration’s tool for assessing risk of bias in randomised trials // BMJ. 2011. V. 343. d5928. https://doi.org/10.1136/bmj.d5928
Реброва О.Ю., Федяева В.К., Хачатрян Г.Р. Адаптация и валидизация вопросника для оценки риска систематических ошибок в рандомизированных контролируемых испытаниях // Мед. технологии. Оценка и выбор. 2015. Т. 1. № 19. С. 9–17.
Wells G., Shea B., O’Connell D. et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses // Ottawa Hospital Res. Institute. 2000. https://www.ohri.ca//programs/clinical_epidemiology/oxford.20asp
Реброва О.Ю., Федяева В.К. Вопросник для оценки риска систематических ошибок в нерандомизированных сравнительных исследованиях: русскоязычная версия шкалы Ньюкасл–Оттава // Мед. технологии. Оценка и выбор. 2016. Т. 3. № 25. С. 14–19.
Campbell H., Rudan I. Interpretation of genetic association studies in complex disease // Pharm. J. 2002. V. 2. P. 349–360. https://doi.org/10.1038/sj.tpj.6500132
Dietrich S., Jacobs S., Zheng J.S. et al. Gene-lifestyle interaction on risk of type 2 diabetes: A systematic review // Obes. Rev. 2019. V. 20. № 11. P. 1557–1571. https://doi.org/10.1111/obr.12921
Nikpay M., Lau P., Soubeyrand S. et al. SGCG rs679482 associates with weight loss success in response to an intensively supervised outpatient program // Diabetes. 2020. V. 69. № 9. P. 2017–2026. https://doi.org/10.2337/db20-0219
Valsesia A., Wang Q.P., Gheldof N. et al. Genome-wide gene-based analyses of weight loss interventions identify a potential role for NKX6.3 in metabolism // Nat. Commun. 2019. V. 10. № 1. P. 540. https://doi.org/10.1038/s41467-019-08492-8
Garaulet M., Smith C.E., Hernández-González T. et al. PPARγ Pro12Ala interacts with fat intake for obesity and weight loss in a behavioural treatment based on the Mediterranean diet // Mol. Nutr. Food Res. 2011. V. 55. № 12. P. 1771–1779. https://doi.org/10.1002/mnfr.201100437
Garaulet M., Vera B., Bonnet-Rubio G. et al. Lunch eating predicts weight-loss effectiveness in carriers of the common allele at PERILIPIN1: the ONTIME (Obesity, Nutrigenetics, Timing, Mediterranean) study // Am. J. Clin. Nutr. 2016. V. 104. № 4. P. 1160–1166. https://doi.org/10.3945/ajcn.116.134528
Garaulet M., Sánchez-Moreno C., Smith C.E. et al. Ghrelin, sleep reduction and evening preference: relationships to CLOCK 3111 T/C SNP and weight loss // PLoS One. 2011. V. 6. № 2. e17435. https://doi.org/10.1371/journal.pone.0017435
Aberle J., Flitsch J., Beck N.A. et al. Genetic variation may influence obesity only under conditions of diet: Analysis of three candidate genes // Mol. Genet. Metab. 2008. V. 95. № 3. P. 188–191. https://doi.org/10.1016/j.ymgme.2008.07.008
Goni L., Riezu-Boj J.I., Milagro F.I. et al. Interaction between an ADCY3 genetic variant and two weight-lowering diets affecting body fatness and body composition outcomes depending on macronutrient distribution: a randomized trial // Nutrients. 2018. V. 10. № 6. P. 789. https://doi.org/10.3390/nu10060789
Ramos-Lopez O., Cuervo M., Goni L. et al. Modeling of an integrative prototype based on genetic, phenotypic, and environmental information for personalized prescription of energy-restricted diets in overweight/obese subjects // Am. J. Clin. Nutr. 2020. V. 111. № 2. P. 459–470. https://doi.org/10.1093/ajcn/nqz286
Garaulet M., Corbalán M.D., Madrid J.A. et al. CLOCK gene is implicated in weight reduction in obese patients participating in a dietary programme based on the Mediterranean diet // Int. J. Obes. (Lond). 2010. V. 34. № 3. P. 516–523. https://doi.org/10.1038/ijo.2009.255
Sakane N., Yoshida T., Umekawa T. et al. Effects of Trp64Arg mutation in the beta 3-adrenergic receptor gene on weight loss, body fat distribution, glycemic control, and insulin resistance in obese type 2 diabetic patients // Diabetes Care. 1997. V. 20. № 12. P. 1887–1890. https://doi.org/10.2337/diacare.20.12.1887
Tchernof A., Starling R.D., Turner A. et al. Impaired capacity to lose visceral adipose tissue during weight reduction in obese postmenopausal women with the Trp64Arg beta3-adrenoceptor gene variant // Diabetes. 2000. V. 49. № 10. P. 1709–1713. https://doi.org/10.2337/diabetes.49.10.1709
Corella D., Qi L., Sorlí J.V. et al. Obese subjects carrying the 11482G>A polymorphism at the perilipin locus are resistant to weight loss after dietary energy restriction // J. Clin. Endocrinol. Metab. 2005. V. 90. № 9. P. 5121–5126. https://doi.org/10.1210/jc.2005-0576
Ruiz J.R., Larrarte E., Margareto J. et al. Preliminary findings on the role of PLIN1 polymorphisms on body composition and energy metabolism response to energy restriction in obese women // Br. J. Nutr. 2011. V. 106. № 4. P. 486–490. https://doi.org/10.1017/S0007114511000432
Qi Q., Bray G.A., Smith S.R. et al. Insulin receptor substrate 1 gene variation modifies insulin resistance response to weight-loss diets in a 2-year randomized trial: the Preventing Overweight Using Novel Dietary Strategies (POUNDS LOST) trial // Circulation. 2011. V. 124. № 5. P. 563–571. https://doi.org/10.1161/CIRCULATIONAHA.111.025767
Ramos-Lopez O., Riezu-Boj J.I., Milagro F.I. et al. Models integrating genetic and lifestyle interactions on two adiposity phenotypes for personalized prescription of energy-restricted diets with different macronutrient distribution // Front. Genet. 2019. V. 10. № 686. https://doi.org/10.3389/fgene.2019.00686
Goni L., Cuervo M., Milagro F.I., Martínez J.A. Gene-Gene interplay and gene-diet interactions involving the MTNR1B rs10830963 variant with body weight loss // J. Nutrigenet. Nutrigenomics. 2014. V. 7. № 4–6. P. 232–242. https://doi.org/10.1159/000380951
de Luis D.A., Izaola O., Primo D., Aller R. Dietary-fat effect of the rs10830963 polymorphism in MTNR1B on insulin resistance in response to 3 months weight-loss diets // Endocrinol. Diabetes Nutr. (Engl. Ed). 2020. V. 67. № 1. P. 43–52. https://doi.org/10.1016/j.endinu.2019.02.007
Chmurzynska A., Muzsik A., Krzyżanowska-Jankowska P. et al. PPARG and FTO polymorphism can modulate the outcomes of a central European diet and a Mediterranean diet in centrally obese postmenopausal women // Nutr. Res. 2019. V. 69. P. 94–100. https://doi.org/10.1016/j.nutres.2019.08.005
Rajkumar A., Lamothe G., Bolongo P. et al. Acyl-CoA synthetase long-chain 5 genotype is associated with body composition changes in response to lifestyle interventions in postmenopausal women with overweight and obesity: A genetic association study on cohorts Montréal–Ottawa New Emerging Team, and Complications Associated with Obesity // BMC Med. Genet. 2016. V. 17. № 1. P. 56. https://doi.org/10.1186/s12881-016-0320-4
Izaola Jáuregui O., López Gómez J.J., Primo Martín D. et al. ACYL-CoA synthetase long-chain 5 polymorphism is associated with weight loss and metabolic changes in response to a partial meal-replacement hypocaloric diet // Nutr. Hosp. 2020. V. 37. № 4. P. 757–762.https://doi.org/10.20960/nh.03019
de Luis D.A., Mulero I., Primo D. et al. Effects of polymorphism rs3123554 in the cannabinoid receptor gene type 2 (CB2R) on metabolic and adiposity parameters after weight loss with two hypocaloric diets // Diabetes Res. Clin. Pract. 2018. V. 139. P. 339–347. https://doi.org/10.1016/j.diabres.2018.02.030
de Luis D., Aller R., Izaola O. et al. Effect of fatty acid-binding protein 2 Ala54Thr genotype on weight loss and cardiovascular risk factors after a high-polyunsaturated fat diet in obese patients // J. Investig. Med. 2012. V. 60. № 8. P. 1194–1198. https://doi.org/10.2310/JIM.0b013e318271fb25
Martinez-Lopez E., Garcia-Garcia M.R., Gonzalez-Avalos J.M. et al. Effect of Ala54Thr polymorphism of FABP2 on anthropometric and biochemical variables in response to a moderate-fat diet // Nutrition. 2013. V. 29. № 1. P. 46–51. https://doi.org/10.1016/j.nut.2012.03.002
de Luis D.A., Aller R., Conde R. et al. The rs9939609 gene variant in FTO modified the metabolic response of weight loss after a 3-month intervention with a hypocaloric diet // J. Investig. Med. 2013. V. 61. № 1. P. 22–26. https://doi.org/10.2310/JIM.0b013e318276161d
Huang T., Qi Q., Li Y. et al. FTO genotype, dietary protein, and change in appetite: the Preventing Overweight Using Novel Dietary Strategies trial // Am. J. Clin. Nutr. 2014. V. 99. № 5. P. 1126–1130. https://doi.org/10.3945/ajcn.113.082164
de Luis D.A., Aller R., Izaola O., Pacheco D. Role of rs9939609 FTO gene variant in weight loss, insulin resistance and metabolic parameters after a high monounsaturated vs a high polyunsaturated fat hypocaloric diets // Nutr. Hosp. 2015. V. 32. № 1. P. 175–181. https://doi.org/10.3305/nh.2015.32.1.9169
de Luis D.A., Aller R., Izaola O. et al. Effects of a high-protein/low-carbohydrate diet versus a standard hypocaloric diet on weight and cardiovascular risk factors: role of a genetic variation in the rs9939609 FTO gene variant // J. Nutrigenet. Nutrigenomics. 2015. V. 8. № 3. P. 128–136. https://doi.org/10.1159/000441142
Verhoef S.P., Camps S.G., Bouwman F.G. et al. Genetic predisposition, dietary restraint and disinhibition in relation to short and long-term weight loss // Physiol. Behav. 2014. V. 128. P. 247–251. https://doi.org/10.1016/j.physbeh.2014.02.004
Holzapfel C., Sag S., Graf-Schindler J. et al. Association between single nucleotide polymorphisms and weight reduction in behavioural interventions-A pooled analysis // Nutrients. 2021. V. 13. № 3. https://doi.org/10.3390/nu13030819
de Luis D.A., Izaola O., Primo D., Aller R. Association of the rs10830963 polymorphism in melatonin receptor type 1B (MTNR1B) with metabolic response after weight loss secondary to a hypocaloric diet based in Mediterranean style // Clin. Nutr. 2018. V. 37. № 5. P. 1563–1568. https://doi.org/10.1016/j.clnu.2017.08.015
de Luis D.A., Izaola O., Primo D., Aller R. A circadian rhythm-related MTNR1B genetic variant (rs10830963) modulate body weight change and insulin resistance after 9 months of a high protein/low carbohydrate vs a standard hypocaloric diet // J. Diabetes Complications. 2020. V. 34. № 4. https://doi.org/10.1016/j.jdiacomp.2020.107534
Goni L., Sun D., Heianza Y., Wang T. et al. A circadian rhythm-related MTNR1B genetic variant modulates the effect of weight-loss diets on changes in adiposity and body composition: the POUNDS Lost trial // Eur. J. Nutr. 2019. V. 58. № 4. P. 1381–1389. https://doi.org/10.1007/s00394-018-1660-y
Grau K., Cauchi S., Holst C. et al. TCF7L2 rs7903146-macronutrient interaction in obese individuals’ responses to a 10-wk randomized hypoenergetic diet // Am J. Clin. Nutr. 2010. V. 91. № 2. P. 472–479. https://doi.org/10.3945/ajcn.2009.27947
Yoon Y., Park B.L., Cha M.H. et al. Effects of genetic polymorphisms of UCP2 and UCP3 on very low calorie diet-induced body fat reduction in Korean female subjects // Biochem. Biophys. Res. Commun. 2007. V. 359. № 3. P. 451–456. https://doi.org/10.1016/j.bbrc.2007.05.110
Stocks T., Angquist L., Banasik K. et al. TFAP2B influences the effect of dietary fat on weight loss under energy restriction // PLoS One. 2012. V. 7. № 8. e43212. https://doi.org/10.1371/journal.pone.0043212
Huang T., Wang T., Heianza Y. et al. HNF1A variant, energy-reduced diets and insulin resistance improvement during weight loss: The POUNDS Lost trial and DIRECT // Diabetes Obes. Metab. 2018. V. 20. № 6. P. 1445–1452. https://doi.org/10.1111/dom.13250
Di Renzo L., Rizzo M., Iacopino L. et al. Body composition phenotype: Italian Mediterranean Diet and C677T MTHFR gene polymorphism interaction // Eur. Rev. Med. Pharmacol. Sci. 2013. V. 17. № 19. P. 2555–2565.
Bojarczuk A., Boulygina E.A., Dzitkowska-Zabielska M. et al. Genome-wide association study of exercise-induced fat loss efficiency // Genes (Basel). 2022. V. 13. № 11. P. 1975. https://doi.org/10.3390/genes13111975
Mazur I.I., Drozdovska S., Andrieieva O. et al. PPARGC1A gene polymorphism is associated with exercise-induced fat loss // Mol. Biol. Rep. 2020. V. 47. № 10. P. 7451–7457. https://doi.org/10.1007/s11033-020-05801-z
Mitchell J.A., Church T.S., Rankinen T. et al. FTO genotype and the weight loss benefits of moderate intensity exercise // Obesity (Silver Spring). 2010. V. 18. № 3. P. 641–643. https://doi.org/10.1038/oby.2009.311
Wang W., Yang K., Wang S. et al. The sex-specific influence of FTO genotype on exercise intervention for weight loss in adult with obesity // Eur. J. Sport Sci. 2022. V. 22. № 12. P. 1926–1931. https://doi.org/10.1080/17461391.2021.1976843
Østergård T., Ek J., Hamid Y. et al. Influence of the PPAR-gamma2 Pro12Ala and ACE I/D polymorphisms on insulin sensitivity and training effects in healthy offspring of type 2 diabetic subjects // Horm. Metab. Res. 2005. V. 37. № 2. P. 99–105. https://doi.org/10.1055/s-2005-861174
Franks P.W., Jablonski K.A., Delahanty L. et al. The Pro12Ala variant at the peroxisome proliferator-activated receptor gamma gene and change in obesity-related traits in the Diabetes Prevention Program // Diabetologia. 2007. V. 50. № 12. P. 2451–2460. https://doi.org/10.1007/s00125-007-0826-6
Orkunoglu-Suer F.E., Gordish-Dressman H., Clarkson P.M. et al. INSIG2 gene polymorphism is associated with increased subcutaneous fat in women and poor response to resistance training in men // BMC Med. Genet. 2008. V. 23. № 9. P. 117. https://doi.org/10.1186/1471-2350-9-117
Cameron J.D., Riou M.È., Tesson F. et al. The TaqIA RFLP is associated with attenuated intervention-induced body weight loss and increased carbohydrate intake in post-menopausal obese women // Appetite. 2013. V. 60. № 1. P. 111–116. https://doi.org/10.1016/j.appet.2012.09.010
Andrade-Mayorga O., Díaz E., Salazar L.A. Effects of four lipid metabolism-related polymorphisms on body composition improvements after 12 weeks of high-intensity interval training and dietary energy restriction in overweight/obese adult women: A pilot study // Front. Physiol. 2021. V. 1. № 12. https://doi.org/10.3389/fphys.2021.712787
Phares D.A., Halverstadt A.A., Shuldiner A.R. et al. Association between body fat response to exercise training and multilocus ADR genotypes // Obes. Res. 2004. V. 12. № 5. P. 807–815. https://doi.org/10.1038/oby.2004.97
de Luis D.A., Gonzalez Sagrado M., Aller R. et al. Influence of the Trp64Arg polymorphism in the beta 3 adrenoreceptor gene on insulin resistance, adipocytokine response, and weight loss secondary to lifestyle modification in obese patients // Eur. J. Intern. Med. 2007. V. 18. № 8. P. 587–592. https://doi.org/10.1016/j.ejim.2007.04.019
Huh J.Y. The role of exercise-induced myokines in regulating metabolism // Arch. Pharm. Res. 2018. V. 41. № 1. P. 14–29. https://doi.org/10.1007/s12272-017-0994-y
Ludwig D.S., Willett W.C., Volek J.S., Neuhouser M.L. Dietary fat: from foe to friend? // Science. 2018. V. 362. P. 764–770.
Wu L., Shen C., Seed Ahmed M. et al. Adenylate cyclase 3: A new target for anti-obesity drug development // Obes. Rev. 2016. V. 17. № 9. P. 907–914. https://doi.org/10.1111/obr.12430
Pitman J.L., Wheeler M.C., Lloyd D.J. et al. A gain-of-function mutation in adenylate cyclase 3 protects mice from diet-induced obesity // PLoS One. 2014. V. 9. № 10. https://doi.org/10.1371/journal.pone.0110226
Tong T., Shen Y., Lee H.W. et al. Adenylyl cyclase 3 haploinsufficiency confers susceptibility to diet-induced obesity and insulin resistance in mice // Sci. Rep. 2016. V. 6. https://doi.org/10.1038/srep34179
URL: https://gtexportal.org.
Markgraf D.F., Al-Hasani H., Lehr S. Lipidomics-reshaping the analysis and perception of type 2 diabetes // Int. J. Mol. Sci. 2016. V. 17. № 11. https://doi.org/10.3390/ijms17111841
Bille D.S., Banasik K., Justesen J.M. et al. Implications of central obesity-related variants in LYPLAL1, NRXN3, MSRA, and TFAP2B on quantitative metabolic traits in adult Danes // PLoS One. 2011. V. 6. № 6. https://doi.org/10.1371/journal.pone.0020640
Kok B.P., Ghimire S., Kim W. et al. Discovery of small-molecule enzyme activators by activity-based protein profiling // Nat. Chem. Biol. 2020. V. 16. № 9. P. 997–1005. https://doi.org/10.1038/s41589-020-0555-4
Lyssenko V., Nagorny C.L., Erdos M.R. et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion // Nat. Genet. 2009. V. 41. № 1. P. 82–88.
Weaver D.R., Reppert S.M. The Mel1a melatonin receptor gene is expressed in human suprachiasmatic nuclei // Neuroreport. 1996. V. 20. № 8. P. 109–112. https://doi.org/10.1097/00001756-199612200-00022
Laermans J., Depoortere I. Chronobesity: Role of the circadian system in the obesity epidemic // Obes. Rev. 2016. V. 17. P. 108–125. https://doi.org/10.1111/obr.12351
Oosterman J.E., Kalsbeek A., la Fleur S.E., Belsham D.D. Impact of nutrients on circadian rhythmicity // Am. J. Physiol. – Regul. Integr. Comp. Physiol. 2015. V. 308. № 5. P. R337–R350. https://doi.org/10.1152/ajpregu.00322.2014
Garaulet M., Madrid J.A. Chronobiology, genetics and metabolic syndrome // Curr. Opin. Lipidol. 2009. V. 20. № 2. P. 127–134. https://doi.org/10.1097/MOL.0b013e3283292399
Naja F., Hasan H., Khadem S.H. et al. Adherence to the Mediterranean diet and its association with sleep quality and chronotype among youth: A cross-sectional study // Front. Nutr. 2022. V. 8. https://doi.org/10.3389/fnut.2021.805955
Corbalán M.D., Morales E.M., Canteras M. et al. Effectiveness of cognitive-behavioral therapy based on the Mediterranean diet for the treatment of obesity // Nutrition. 2009. V. 25. № 7–8. P. 861–869. https://doi.org/10.1016/j.nut.2009.02.013
Fu J., Tan L.J., Lee J.E., Shin S. Association between the mediterranean diet and cognitive health among healthy adults: A systematic review and meta-analysis // Front. Nutr. 2022. V. 9. https://doi.org/10.3389/fnut.2022.946361
Maciejewska-Skrendo A., Massidda M., Tocco F., Leźnicka K. The influence of the differentiation of genes encoding peroxisome proliferator-activated receptors and their coactivators on nutrient and energy metabolism // Nutrients. 2022. V. 14. № 24. https://doi.org/10.3390/nu14245378
Kubota N., Terauchi Y., Miki H. et al. PPAR gamma mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance // Mol. Cell. 1999. V. 4. № 4. P. 597–609. https://doi.org/10.1016/s1097-2765(00)80210-5
Marcinkiewicz A., Gauthier D., Garcia A. et al. The phosphorylation of serine 492 of perilipin a directs lipid droplet fragmentation and dispersion // J. Biol. Chem. 2006. V. 281. P. 11901–11909.
Rankinen T., Rice T., Teran-Garcia M. et al. FTO genotype is associated with exercise training-induced changes in body composition // Obesity (Silver Spring). 2010. V. 18. № 2. P. 322–326. https://doi.org/10.1038/oby.2009.205
Danaher J., Stathis C.G., Wilson R.A. et al. High intensity exercise downregulates FTO mRNA expression during the early stages of recovery in young males and females // Nutr. Metab. (Lond). 2020. V. 17. № 68. https://doi.org/10.1186/s12986-020-00489-1
Wu W., Feng J., Jiang D. et al. AMPK regulates lipid accumulation in skeletal muscle cells through FTO-dependent demethylation of N6-methyladenosine // Sci. Rep. 2017. V. 7. https://doi.org/10.1038/srep41606
Li S., Zhao J.H., Luan J. et al. Physical activity attenuates the genetic predisposition to obesity in 20,000 men and women from EPIC-Norfolk prospective population study // PLoS Med. 2010. V. 7. № 8. https://doi.org/10.1371/journal.pmed.1000332
Loos R.J., Yeo G.S. The bigger picture of FTO: The first GWAS-identified obesity gene // Nat. Rev. Endocrinol. 2014. V. 10. № 1. P. 51–61. https://doi.org/10.1038/nrendo.2013.227
Kilpeläinen T.O., Qi L., Brage S. et al. Physical activity attenuates the influence of FTO variants on obesity risk: A meta-analysis of 218 166 adults and 19 268 children // PLoS Med. 2011. V. 8. № 11. https://doi.org/10.1371/journal.pmed.1001116
Vimaleswaran K.S., Li S., Zhao J.H. et al. Physical activity attenuates the body mass index-increasing influence of genetic variation in the FTO gene // Am. J. Clin. Nutr. 2009. V. 90. № 2. P. 425–428. https://doi.org/10.3945/ajcn.2009.27652
Reddon H., Gerstein H.C., Engert J.C. et al. Physical activity and genetic predisposition to obesity in a multiethnic longitudinal study // Sci. Rep. 2016. V. 6. https://doi.org/10.1038/srep18672
Zarebska A., Jastrzebski Z., Cieszczyk P. et al. The Pro12Ala polymorphism of the peroxisome proliferator-activated receptor gamma gene modifies the association of physical activity and body mass changes in Polish Women // PPAR Res. 2014. V. 2014. https://doi.org/10.1155/2014/373782
Li S., He C., Nie H. et al. G allele of the rs1801282 polymorphism in PPARγ gene confers an increased risk of obesity and hypercholesterolemia, While T allele of the rs3856806 polymorphism displays a protective role against dyslipidemia: A systematic review and meta-analysis // Front. Endocrinol. (Lausanne). 2022. V. 13. https://doi.org/10.3389/fendo.2022.919087
Masud S., Ye S., SAS Group. Effect of the peroxisome proliferator activated receptor-gamma gene Pro12Ala variant on body mass index: a meta-analysis // J. Med. Genet. 2003. V. 40. № 10. https://doi.org/10.1136/jmg.40.10.773
Qi Q., Chu A.Y., Kang J.H. et al. Sugar-sweetened beverages and genetic risk of obesity // N. Engl. J. Med. 2012. V. 367. P. 1387–1396. https://doi.org/10.1056/NEJMoa1203039
Qi Q., Chu A.Y., Kang J.H. et al. Fried food consumption, genetic risk, and body mass index: Gene-diet interaction analysis in three us cohort studies // BMJ. 2014. V. 348. https://doi.org/10.1136/bmj.g1610
Jääskeläinen A., Schwab U., Kolehmainen M. et al. Meal frequencies modify the effect of common genetic variants on body mass index in adolescents of the northern Finland birth cohort 1986 // PLoS One. 2013. V. 10. № 9. e73802. https://doi.org/10.1371/journal.pone.0073802
Cienfuegos S., Gabel K., Kalam F. et al. Effects of 4- and 6-h time-restricted feeding on weight and cardiometabolic health: A randomized controlled trial in adults with obesity // Cell Metab. 2020. V. 32. № 3. P. 366–378. https://doi.org/10.1016/j.cmet.2020.06.018
Pellegrini M., Cioffi I., Evangelista A. et al. Effects of time-restricted feeding on body weight and metabolism. A systematic review and meta-analysis // Rev. Endocr. Metab. Disord. 2020. V. 21. № 1. P. 17–33. https://doi.org/10.1007/s11154-019-09524-w
Heianza Y., Sun D., Wang T. et al. Starch digestion-related amylase genetic variant affects 2-year changes in adiposity in response to weight-loss diets: The POUNDS Lost Trial // Diabetes. 2017. V. 66. № 9. P. 2416–2423. https://doi.org/10.2337/db16-1482
Zhang X., Qi Q., Zhang C. et al. FTO genotype and 2-year change in body composition and fat distribution in response to weight-loss diets: the POUNDS LOST Trial // Diabetes. 2013. V. 62. № 2. P. 662. https://doi.org/10.2337/db11-1799
Frey U.H., Hauner H., Jöckel K.H. et al. A novel promoter polymorphism in the human gene GNAS affects binding of transcription factor upstream stimulatory factor 1, Galphas protein expression and body weight regulation // Pharmacogenet. Genomics. 2008. V. 18. № 2. P. 141–151. https://doi.org/10.1097/FPC.0b013e3282f49964
Razquin C., Martinez J.A., Martinez-Gonzalez M.A. et al. A Mediterranean diet rich in virgin olive oil may reverse the effects of the -174G/C IL6 gene variant on 3-year body weight change // Mol. Nutr. Food Res. 2010. V. 54. Suppl. 1. P. S75–S82. https://doi.org/10.1002/mnfr.200900257
Heianza Y., Sun D., Ma W. et al. Gut-microbiome-related LCT genotype and 2-year changes in body composition and fat distribution: the POUNDS Lost Trial // Int. J. Obes. (Lond). 2018. V. 42. № 9. P. 1565–1573. https://doi.org/10.1038/s41366-018-0046-9
Sun D., Heianza Y., Li X. et al. Genetic, epigenetic and transcriptional variations at NFATC2IP locus with weight loss in response to diet interventions: The POUNDS Lost Trial // Diabetes Obes. Metab. 2018. V. 20. № 9. P. 2298–2303. https://doi.org/10.1111/dom.13333
Lin X., Qi Q., Zheng Y. et al. Neuropeptide Y genotype, central obesity, and abdominal fat distribution: the POUNDS Lost Trial // Am. J. Clin. Nutr. 2015. V. 102. № 2. P. 514–519. https://doi.org/10.3945/ajcn.115.107276
Valeeva F.V., Medvedeva M.S., Khasanova K.B. et al. Association of gene polymorphisms with body weight changes in prediabetic patients // Mol. Biol. Rep. 2022. V. 49. № 6. P. 4217–4224. https://doi.org/10.1007/s11033-022-07254-y
Heianza Y., Ma W., Huang T. et al. macronutrient intake-associated FGF21 genotype modifies effects of weight-loss diets on 2-year changes of central adiposity and body composition: The POUNDS Lost Trial // Diabetes Care. 2016. V. 39. № 11. P. 1909–1914. https://doi.org/10.2337/dc16-1111
Xu M., Qi Q., Liang J. et al. Genetic determinant for amino acid metabolites and changes in body weight and insulin resistance in response to weight-loss diets: the Preventing Overweight Using Novel Dietary Strategies (POUNDS LOST) trial // Circulation. 2013. V. 127. № 12. P. 1283–1289. https://doi.org/10.1161/CIRCULATIONAHA.112.000586
Mansego M.L., Milagro F.I., Zulet M.A., Martinez J.A. SH2B1 CpG-SNP is associated with body weight reduction in obese subjects following a dietary restriction program // Ann. Nutr. Metab. 2015. V. 66. № 1. P. 1–9. https://doi.org/10.1159/000368425
Mattei J., Qi Q., Hu F.B. et al. TCF7L2 genetic variants modulate the effect of dietary fat intake on changes in body composition during a weight-loss intervention // Am. J. Clin. Nutr. 2012. V. 96. № 5. P. 1129–1136. https://doi.org/10.3945/ajcn.112.038125
Aberle J., Evans D., Beil F.U., Seedorf U. A polymorphism in the apolipoprotein A5 gene is associated with weight loss after short-term diet // Clin. Genet. 2005. V. 68. № 2. P. 152–154. https://doi.org/10.1111/j.1399-0004.2005.00463.x
Hamada T., Kotani K., Nagai N. et al. Genetic polymorphisms of the renin-angiotensin system and obesity-related metabolic changes in response to low-energy diets in obese women // Nutrition. 2011. V. 27. № 1. P. 34–39. https://doi.org/10.1016/j.nut.2009.10.012
Tsuzaki K., Kotani K., Nagai N. et al. Adiponectin gene single-nucleotide polymorphisms and treatment response to obesity // J. Endocrinol. Invest. 2009. V. 32. № 5. P. 395–400. https://doi.org/10.1007/BF03346474
Ruiz J.R., Larrarte E., Margareto J. et al. Role of β2-adrenergic receptor polymorphisms on body weight and body composition response to energy restriction in obese women: preliminary results // Obesity (Silver Spring). 2011. V. 19. № 1. P. 212–215. https://doi.org/10.1038/oby.2010.130
de Luis D.A., Fernández Ovalle H., Izaola O. et al. Rs 10767664 gene variant in Brain Derived Neurotrophic Factor (BDNF) affect metabolic changes and insulin resistance after a standard hypocaloric diet // J. Diabetes Complications. 2018. V. 32. № 2. P. 216–220. https://doi.org/10.1016/j.jdiacomp.2017.10.005
de Luis D.A., Gonzalez Sagrado M., Aller R. et al. Effects of C358A missense polymorphism of the endocannabinoid degrading enzyme fatty acid amide hydrolase on weight loss after a hypocaloric diet // Metabolism. 2011. V. 60. № 5. P. 730–734. https://doi.org/10.1016/j.metabol.2010.07.007
Mammès O., Aubert R., Betoulle D. et al. LEPR gene polymorphisms: associations with overweight, fat mass and response to diet in women // Eur. J. Clin. Invest. 2001. V. 31. № 5. P. 398–404. https://doi.org/10.1046/j.1365-2362.2001.00843.x
Abete I., Goyenechea E., Crujeiras A.B., Martínez J.A. Inflammatory state and stress condition in weight-lowering Lys109Arg LEPR gene polymorphism carriers // Arch. Med. Res. 2009. V. 40. № 4. P. 306–310. https://doi.org/10.1016/j.arcmed.2009.03.005
Thamer C., Machann J., Stefan N. et al. Variations in PPARD determine the change in body composition during lifestyle intervention: A whole-body magnetic resonance study // J. Clin. Endocrinol. Metab. 2008. V. 93. № 4. P. 1497–1500. https://doi.org/10.1210/jc.2007-1209
Matsuo T., Nakata Y., Katayama Y. et al. PPARG genotype accounts for part of individual variation in body weight reduction in response to calorie restriction // Obesity (Silver Spring). 2009. V. 17. № 10. P. 1924–1931. https://doi.org/10.1038/oby.2009.199
Yamakage H., Konishi Y., Muranaka K. et al. Association of protein tyrosine phosphatase 1B gene polymorphism with the effects of weight reduction therapy on bodyweight and glycolipid profiles in obese patients // J. Diabetes Investig. 2021. V. 12. № 8. P. 1462–1470. https://doi.org/10.1111/jdi.13492
Heni M., Herzberg-Schäfer S., Machicao F. et al. Dietary fiber intake modulates the association between variants in TCF7L2 and weight loss during a lifestyle intervention // Diabetes Care. 2012. V. 35. № 3. e24. https://doi.org/10.2337/dc11-2012
Nagai N., Sakane N., Kotani K. et al. Uncoupling protein 1 gene —3826 A/G polymorphism is associated with weight loss on a short-term, controlled-energy diet in young women // Nutr. Res. 2011. V. 31. № 4. P. 255–261. https://doi.org/10.1016/j.nutres.2011.03.010
Cha M.H., Kim K.S., Suh D., Yoon Y. Effects of genetic polymorphism of uncoupling protein 2 on body fat and calorie restriction-induced changes // Hereditas. 2007. V. 144. № 5. P. 222–227. https://doi.org/10.1111/j.2007.0018-0661.02005.x
Papazoglou D., Papathanasiou P., Papanas N. et al. Uncoupling protein-2 45-base pair insertion/deletion polymorphism: is there an association with severe obesity and weight loss in morbidly obese subjects? // Metab. Syndr. Relat. Disord. 2012. V. 10. № 4. P. 307–311. https://doi.org/10.1089/met.2012.0003
de Luis D.A., Aller R., Izaola O. et al. Relation of ‒55CT polymorphism of UCP3 gene with weight loss and metabolic changes after a high monounsaturated fat diet in obese non diabetic patients // Eur. Rev. Med. Pharmacol. Sci. 2013. V. 17. № 20. P. 2810–2815.
Cha M.H., Shin H.D., Kim K.S. et al. The effects of uncoupling protein 3 haplotypes on obesity phenotypes and very low-energy diet-induced changes among overweight Korean female subjects // Metabolism. 2006. V. 55. № 5. P. 578–586. https://doi.org/10.1016/j.metabol.2005.11.012
Corbi G., Polito R., Monaco M.L. et al. Adiponectin expression and genotypes in Italian people with severe obesity undergone a hypocaloric diet and physical exercise program // Nutrients. 2019. V. 11. № 9. https://doi.org/10.3390/nu11092195
Leońska-Duniec A., Jastrzębski Z., Jażdżewska A. et al. Individual responsiveness to exercise-induced fat loss and improvement of metabolic profile in young women is associated with polymorphisms of adrenergic receptor genes // J. Sports Sci. Med. 2018. V. 17. № 1. P. 134–144.
Suchanek P., Kralova-Lesna I., Poledne R. et al. An AHSG gene variant modulates basal metabolic rate and body composition development after a short-time lifestyle intervention // Neuro Endocrinol. Lett. 2011. V. 32. Suppl. 2. P. 32–36.
Tworoger S.S., Chubak J., Aiello E.J. et al. The effect of CYP19 and COMT polymorphisms on exercise-induced fat loss in postmenopausal women // Obes. Res. 2004. V. 12. № 6. P. 972–981. https://doi.org/10.1038/oby.2004.119
de Luis D.A., Aller R., Izaola O. et al. Influence of ALA54THR polymorphism of fatty acid binding protein 2 on lifestyle modification response in obese subjects // Ann. Nutr. Metab. 2006. V. 50. № 4. P. 354–360. https://doi.org/10.1159/000094299
Franzago M., Di Nicola M., Fraticelli F. et al. Nutrigenetic variants and response to diet/lifestyle intervention in obese subjects: A pilot study // Acta Diabetol. 2022. V. 59. № 1. P. 69–81. https://doi.org/10.1007/s00592-021-01787-7
Ficek K., Ciȩszczyk P., Leźnicka K. et al. Novel associations between interleukin-15 polymorphisms and post-training changes of body composition parameters in young nonobese women // Front. Physiol. 2019. V. 5. № 10. https://doi.org/10.3389/fphys.2019.00876
Suchánek P., Lánská V., Hubáček J.A. Body composition changes in adult females after lifestyle intervention are influenced by the NYD-SP18 variant // Cent. Eur. J. Publ. Health. 2015. V. 23. Suppl. Р. 19–S22. https://doi.org/10.21101/cejph.a4105
Leońska-Duniec A., Cieszczyk P., Jastrzębski Z. et al. The polymorphisms of the PPARD gene modify post-training body mass and biochemical parameter changes in women // PLoS One. 2018. V. 13. № 8. https://doi.org/10.1371/journal.pone.0202557
Haupt A., Thamer C., Heni M. et al. Gene variants of TCF7L2 influence weight loss and body composition during lifestyle intervention in a population at risk for type 2 diabetes // Diabetes. 2010. V. 59. № 3. P. 747–750. https://doi.org/10.2337/db09-1050
Lim K.I., Shin Y.A. Impact of UCP2 polymorphism on long-term exercise-mediated changes in adipocytokines and markers of metabolic syndrome // Aging Clin. Exp. Res. 2014. V. 26. № 5. P. 491–496. https://doi.org/10.1007/s40520-014-0213-3
de Luis D.A., Aller R., Izaola O. et al. Modulation of adipocytokines response and weight loss secondary to a hypocaloric diet in obese patients by –55CT polymorphism of UCP3 gene // Horm. Metab. Res. 2008. V. 40. № 3. P. 214–218. https://doi.org/10.1055/s-2008-1046796
Дополнительные материалы
- скачать ESM.docx
- Приложение 1.
Таблица 1. Оценка уровней риска систематической ошибки в РКИ
Таблица 2. Оценка риска систематических ошибок нерандомизированных контролируемых исследований и итоговые оценки риска в соответствии со шкалой Ньюкасла-Оттавы
Таблица 3. Оценка методологического качества исследования в соответствии с критериями, важными для исследований, изучающих генетическую ассоциацию
Таблица 4. Оценка методологического качества исследования в соответствии с критериями, важными для исследований, изучающих генетическую ассоциацию
Таблица 5. Полиморфизмы генов, ассоциированные с эффективностью диетотерапии
Таблица 6. Полиморфизмы генов, ассоциированные с эффективностью физической нагрузки