Известия РАН. Серия биологическая, 2021, № 1, стр. 30-43
Коронавирусные инфекции животных: будущие риски для человека
И. М. Донник 1, Иг. В. Попов 2, 3, С. В. Середа 2, Ил. В. Попов 3, М. Л. Чикиндас 2, 4, А. М. Ермаков 2, *
1 Российская академия наук
119991 Москва, Ленинский просп., 14, Россия
2 Донской государственный технический университет
344000 Ростов-на-Дону, пл. Гагарина, 1, Россия
3 Ростовский государственный медицинский университет
344022 Ростов-на-Дону, пер. Нахичеванский, 29, Россия
4 Health Promoting Naturals Laboratory, School of Environmental and Biological Sciences, Rutgers, the State University of New Jersey
NJ 08901 New Brunswick, 65 Dudley Road, USA
* E-mail: amermakov@yandex.ru
Поступила в редакцию 04.05.2020
После доработки 27.06.2020
Принята к публикации 17.07.2020
Аннотация
Установлено, что коронавирусы обладают огромным эволюционным потенциалом, и на данный момент в новейшей истории человечества отмечены три крупные вспышки новых коронавирусных инфекций человека. Определены закономерности возникновения новых зоонозных коронавирусных инфекций и роль биоветеринарного контроля в предотвращении их потенциальных вспышек в будущем, а также рассмотрена возможность заражения животных-компаньонов SARS-CoV-2. Продемонстрировано, что вмешательство человека в дикую природу приводит к столкновению в одной пространственно-временнóй точке разных видов животных и их вирусов, провоцируя подчас появление новых, непредсказуемых форм патогенных вирусов. Рассмотрена возможность использования пробиотиков для контроля вирусных инфекций у животных.
Коронавирусы (Coronaviridae (CoV)) – семейство патогенных вирусов, поражающих человека и животных и вызывающих заболевания дыхательной, желудочно-кишечной и нервной систем различной степени тяжести. Семейство CoV включает в себя два подсемейства: Coronavirinae, которое подразделяется на четыре рода (Alphacoronavirus, Betacoronavirus, Gammacoronavirus и Deltacoronavirus), и Torovirinae, состоящее из двух родов (Torovirus и Bafinivirus). Род Betacoronavirus подразделяется на несколько подродов, которые до пересмотра таксономии в 2019 г. носили названия согласно первым четырем буквам латинского алфавита. В настоящее время принято считать, что этот род состоит из подродов Embecovirus (бывший подрод A), Sarbecovirus (бывший подрод B), Merbecovirus (бывший подрод C), Nobecovirus (бывший подрод D) и впервые отдельно выделенный подрод Hibecovirus (Li et al., 2019). Благодаря уникальному механизму репликации CoV наблюдается высокая частота возникновений генетических рекомбинаций в их РНК и последующих мутаций, что в совокупности обеспечивает CoV способностью к быстрой адаптации к новым хозяевам и экологическим нишам (Ji et al., 2020; Lu et al., 2020).
Мутационная активность способствует изменению вирулентности CoV, однако не последнюю роль в этом играют и животные-носители, которые вовлечены в глобальную экосистему, в том числе и в качестве основополагающих звеньев зоонозных вирусных инфекций – первичных и вторичных резервуаров (Omrani et al., 2015; Mohd et al., 2016; Shi et al., 2017). На данный момент, исходя из ретроспективных эпидемиологических данных, с уверенностью можно утверждать, что наибольшим эмерджентным зоонозным потенциалом обладают бета-коронавирусы, такие как SARS-CoV, MERS-CoV и SARS-CoV-2. Следует отметить, что к возникновению и распространению этих вирусов причастны как домашние, так и дикие животные, в частности летучие мыши (Sun et al., 2020). Особенности антивирусных иммунных реакций этих животных создали удобный фундамент для более интенсивного развития прародителей трех вышеперечисленных вирусов (Li et al., 2019). Сейчас перед научным сообществом стоит задача разработки стратегии предотвращения потенциальной четвертой вспышки новой коронавирусной инфекции.
МАТЕРИАЛЫ И МЕТОДЫ
Для написания этого систематического обзора нами был проведен поиск данных в базах Scopus, The Cochrane Database, MEDLINE/PubMed Database, Embase-Elsevier, Web of Science Core Collection, eLIBRARY (2003–2020 гг.) c использованием комбинаций ключевых слов и логического оператора SQL: “coronavirus” AND (“animal” OR “bat” OR “dromedary camel” OR “civet” OR “pangolin” OR “cat” OR “dog” OR “ferret” OR “animal model”). Мы воспользовались информацией, представленной во многих (91) крупных обзорах, а также результатами систематических анализов и научно-исследовательских работ, в которых были обсуждены фундаментальные и клинические аспекты коронавирусных инфекций различных видов животных и их зоонозный потенциал.
РЕЗУЛЬТАТЫ И ОБСУЖДЕНИЕ
Установлено, что все известные человеческие коронавирусные инфекции имеют зоонозную природу. В систематическом обзоре (Ye et al., 2020) приведено семь примеров выявленных человеческих CoV, в формировании которых задействованы не менее пяти представителей разных отрядов млекопитающих.
Животное рассматривается как первичный эволюционный хозяин человеческого CoV, если у него обнаружен вирус-предшественник, гомологичный человеческому на уровне нуклеотидной последовательности. Этот вирус обычно не имеет выраженной вирулентности в отношении животного-хозяина, и его носительство протекает бессимптомно. В большинстве случаев, для того чтобы инфицировать человека, CoV должны преодолеть межвидовой барьер, что обеспечивается разнообразием генетических и фенотипических качеств вируса, возникающих из-за ошибок во время репликации, например при инактивации экзорибонуклеазы (Ogando et al., 2019). Патогенность вируса-предшественника часто проявляется при инфицировании промежуточного хозяина, который играет роль резервуара для интенсивного размножения. При повторяющемся контакте с зараженным животным вирус-предшественник имеет большие шансы перейти к человеку, причем если CoV способны к трансмиссии между людьми, они не теряют возможности к развитию и могут адаптироваться к организму нового хозяина с потерей вирулентных качеств. Так, MERS-CoV обладает самой высокой летальностью среди семи выявленных CoV. При этом MERS-CoV обладает меньшей контагиозностью среди людей по сравнению с другими представителями этой группы вирусов (Omrani et al., 2015; Chu et al., 2020). В то же время HCoV-229E, HCoV-OC43, HCoV-NL63 и HCoV-HKU1 не вызывают серьезных и системных воспалительных заболеваний у человека, что свидетельствует об адаптационных свойствах этих вирусов к новым хозяевам (Corman et al., 2015; Milewska et al., 2018; Cui et al., 2019). Интересна ситуация с SARS-CoV-2: в ходе эволюции контагиозные и вирулентные качества этого вируса сформировались наиболее благоприятно для его пандемического распространения. Однако при рассмотрении этого вируса прослеживается логика адаптационного превалирования контагиозности над вирулентностью. Танг с соавт. обнаружили два типа SARS-CoV-2 (более вирулентный L-тип и менее вирулентный S-тип) и выяснили, что в условиях санитарно-эпидемиологических мер по борьбе с распространением этого вируса в Китае трансмиссия S-типа, вызывающего относительно менее тяжелые формы воспалительных заболеваний, начала превалировать над L-типом (Tang et al., 2020).
Таким образом, для обнаружения потенциальных эмерджентных зоонозных коронавирусных инфекций необходимо рассматривать варианты их бессимптомного носительства у животных, особенно если инфекционный агент – представитель рода Betacoronavirus.
Представители отряда Chiroptera (рукокрылые) причастны к возникновению множества эпидемиологических вспышек зоонозных вирусных инфекций, в частности общих для других животных и людей. Среди человеческих вирусов, предшественники которых замечены у этих животных, выделяют филовирусы (Эбола и Марбург) (Goldstein et al., 2018; Forbes et al., 2019), парамиксовирусы (Нипах, Хендра) (Drexler et al., 2012) и, конечно, CoV (Ye et al., 2020). Кроме того, необходимо отметить, что сегодня установлено не менее 30 видов CoV, для которых рукокрылые – природный резервуар (Wong et al., 2019). Один из благоприятных факторов для существования столь большого числа зоонозных вирусов – видовое разнообразие отряда рукокрылых (>1300 видов), а значит, и большое число разных клеток и рецепторов, с которыми могут взаимодействовать инфекционные агенты. Немаловажную роль в обширной межвидовой трансмиссии зоонозных вирусов играет и факт того, что рукокрылые – единственные млекопитающие, способные на длительные перелеты, что повышает число контактов с другими животными (Hawkins et al., 2019).
Однако главная причина того, что эти животные считаются идеальными “инкубаторами” новых патогенных вирусов, в том числе и CoV, – бессимптомное вирусоносительство, обусловленное ограниченными ответными иммунно-воспалительными реакциями. При попадании вирусов в организм у большинства млекопитающих активируются провоспалительные цитокины, что приводит к воспалительным заболеваниям и летальном исходам (Tseng et al., 2012; Collins et al., 2014). Недавнее исследование (Brook et al., 2020) продемонстрировало, что при взаимодействии клеток крыланов (Pteropodidae) in vitro с разными подтипами везикуловирусов Индианы наблюдается довольно быстрый иммунный ответ, который в то же время индуцирует высокочастотную репликацию вируса в культуре клеток. Кроме того, ученые также обнаружили, что антивирусный интерферонопосредованный ответ летучих мышей не вызывает воспалительной реакции, достаточной для повреждения клеток, что в свою очередь продлевает предполагаемое персистирование вирусной инфекции. Подобные результаты подтверждаются рядом ранее проведенных исследований. Так, было обнаружено, что у рукокрылых сигнальные пути NF-κB дегенерированы (Zhang et al., 2013), а активность NLRP3-инфламасом подавлена по сравнению с таковыми у других животных (Ahn et al., 2019). Этим обосновывается ограниченность воспалительного ответа и бессимптомное течение вирусных инфекций. Было показано также, что повышенная регуляция NK-клеток NKG2/CD94 и низкая экспрессия основных молекул комплекса гистосовместимости класса I у крыланов могут препятствовать работе NK-клеток (Pavlovich et al., 2018).
Все эти особенности иммунитета делают организм рукокрылых идеальной средой для развития и репликации большого числа вирусов, в частности CoV. Имеются также данные, что активные формы кислорода, образующиеся вследствие активного метаболизма этих млекопитающих, могут оказывать дополнительное мутагенное влияние на CoV посредством воздействия на РНК-зависимую РНК-полимеразу (Zhang et al., 2013).
Таким образом, рукокрылые представляют опасность как первичное звено в возникновении эмерджентных коронавирусных инфекций. CoV, которые приобрели высокую репликационную активность за счет длительного персистирования в организме рукокрылых, при межвидовой трансмиссии имеют высокие шансы приобретения вирулентных качеств по отношению к новым хозяевам, что, вероятно, и произошло при эволюционном развитии SARS-CoV, MERS-CoV и SARS-CoV-2.
Прямая передача CoV от рукокрылых человеку маловероятна из-за низкой частоты контактов с этими животными. Поэтому в большинстве случаев человеческие CoV при межвидовой трансмиссии преодолели этап другого животного-переносчика как промежуточного хозяина.
Первая вспышка человеческой коронавирусной инфекции была вызвана SARS-CoV в ноябре 2002 г. в провинции Гуандун, Китай (Perlman et al., 2009). Вспышка инициировала обширное исследование, целью которого было установление источника инфекции. В 2003 г. у работников животного рынка Шэньчжэня и у животных, в частности у гималайских цивет Paguma larvata и енотовидных собак Nyctereutes procyonoides, был выявлен SARS-подобный CoV, который был на 99.8% гомологичен SARS-CoV (Guan et al., 2003). При экспериментальном моделировании коронавирусной инфекции циветы оказались восприимчивы и к SARS-CoV, и к SARS-подобному CoV, после чего именно они стали считаться животным резервуаром (Wu et al., 2005). Немного позже был обнаружен предположительный CoV-предшественник, на 95% идентичный по нуклеотидной последовательности SARS-CoV у китайских рыжих подковоносых летучих мышей Rhinolophus sinicus (Lau et al., 2005; Li et al., 2005), что окончательно установило цепочку вирусной трансмиссии и эволюции.
На примере этой вспышки мы считаем нужным отметить несколько важных фактов. Во-первых, известно, что рукокрылых до выявления у них вероятного предшественника SARS-CoV никто не рассматривал как резервуар зоонозных вирусных инфекций (Wong et al., 2019). Это свидетельствует о том, что человечество может обнаружить других представителей животного мира, организм которых также может оказаться идеальной средой для “инкубации” новых эмерджентных вирусных инфекций. Во-вторых, антитела к SARS-подобным CoV идентифицировали только у гималайских цивет, которые находились на рынке диких животных, у домашних и диких представителей этого вида антител обнаружено не было (Tu et al., 2004). Это может свидетельствовать об отсутствии необходимости большого числа особей для участия в межвидовой трансмиссии CoV в качестве промежуточного звена. В-третьих, у всего семейства виверровых (Viverridae), к которым относятся циветы, согласно недавнему систематическому обзору (Wicker et al., 2017) достоверно установлены десять вирусных инфекций – только одна из них имеет коронавирусную природу. Следовательно, при возникновении новых эмерджентных вирусных инфекций с эпидемическим потенциалом промежуточным звеном может быть любое животное, находящееся в тесном повторяющемся контакте с человеком. Наличие коронавирусной инфекции в видовом анамнезе не обязательно.
Вторая вспышка ранее неизвестной коронавирусной инфекции, вызванная MERS-CoV, имеет некоторые отличия от вышеописанного сценария трансмиссии и развития SARS-CoV. Впервые вирус идентифицировали в 2012 г. в Саудовской Аравии (Milne-Price et al., 2014), позже были установлены генетически сходные CoV рукокрылых – CoV-HKU4, CoV-HKU5 и CoV-HKU25, которые на 75–87% гомологичны MERS-CoV. Все эти вирусы для клеточной инвазии используют дипептидилпептидазу-4 (DPP4) в качестве рецептора (van Boheemen et al., 2012; Lau et al., 2018; Luo et al., 2018). Однако в отличие от SARS-CoV у животного (предположительно природного резервуара), а именно одногорбого верблюда Camelus dromedarius, был идентифицирован MERS-CoV, на 100% гомологичный тому, который выделили у людей (Raj et al., 2014a). Это свидетельствует о том, что MERS-CoV эволюционно дальше от CoV рукокрылых и момент от трансмиссии потенциального CoV-предшественника верблюдам от рукокрылых до формирования MERS-CoV более длительный. Кроме того, на потенциальное происхождение MERS-CoV от CoV рукокрылых указывает фекально-оральный путь передачи, в большей степени характерный для последних (Samara, Abdoun, 2014).
В отличие от коронавирусных инфекций цивет таковые у верблюдовых (Camelidae) встречаются довольно часто, причем некоторые из них даже способны преодолеть межвидовой барьер и перейти к человеку, тем самым приобретя зоонозную природу. Так, в 2018 г. Корман с соавт. опубликовали данные исследования, согласно которым HCoV-229E, вызывающий нелетальные инфекции дыхательных путей, может передаваться от верблюдов людям путем, аналогичным путям трансмиссии MERS-CoV. Это свидетельствует о важной роли верблюдов в качестве резервуара коронавирусных инфекций (Corman et al., 2018). В 2014 г. Ву с соавт. сообщили об идентификации у одногорбых верблюдов Средней Азии нового CoV из рода Betacoronavirus – DcCoV UAE-HKU23, а в 2016 г. антитела против этого CoV были установлены у 98.3–100% верблюдов этого же региона (Woo et al., 2014, 2016). В 2019 г. была опубликована работа о генетическом разнообразии разных штаммов UAE-HKU23, в которой авторы предположили, что эволюционное развитие этого CoV, основанное на приобретении новых генетических качеств в процессе репликации, идет по пути MERS-CoV, что свидетельствует о его возрастающем эмерджентном потенциале (So et al., 2019).
Верблюдовые – идеальный промежуточный хозяин зоонозных инфекций, поскольку в некоторых регионах они находятся в тесных и повторяющихся контактах с людьми, используются для производства молока и мяса и в качестве ездовых животных (Widagdo et al., 2019). Также одногорбые верблюды являются резервуаром 37 зоонозных инфекций, 13 из которых имеют вирусную природу (Zhu et al., 2019).
Одногорбые верблюды могут быть носителями коронавирусной инфекции, которая встречается у других представителей семейства верблюдовых. На основании 1309 образцов ректальных и назальных мазков, отобранных у верблюдов в Саудовской Аравии между маем 2014 г. и апрелем 2015 г., были обнаружены два представителя рода Betacoronavirus (MERS-CoV и HKU23-CoV) и один представитель рода Alphacoronavirus, который оказался генетически гомологичен CoV, выявленному у альпак Vicugna pacos в США в 2007 г., и HCoV-229E человека (Sabir et al., 2016). Позже с помощью секвенирования полного генома выявленного альфа-CoV обнаружилось, что этот вирус на 92.2% гомологичен HCoV-229, а общий предок этих двух вирусов мог перейти от альпак к людям в 1960-х гг. Этот факт установлен благодаря сравнению различных штаммов альфа-CoV, идентифицированных в промежутке между 1962 и 2003 гг. (Crossley et al., 2010, 2012).
Верблюдовые не единственные сельскохозяйственные животные, которые вовлечены в трансмиссионные цепочки CoV, так как все жвачные сельскохозяйственные животные могут быть инфицированы CoV, что наносит экономический ущерб глобальному животноводству, а некоторые CoV этих животных рассматриваются в качестве предшественников человеческих вирусов. Распространению CoV среди скота способствуют такие факторы, как плотное содержание поголовья на ограниченной территории, благоприятное условие для быстрой трансмиссии вирусов фекально-оральным и воздушно-капельным путями (Heckert et al., 1990; Decaro et al., 2008). Достоверно известно, что зимой иммунная резистентность животных снижается, а это сказывается на повышенной частоте заболеваемости, обусловленной коронавирусными инфекциями (Carman et al., 1992). В совокупности это также является подходящей средой для формирования потенциальных зоонозных CoV. Вийген с соавт. предположили, что HCoV-OC43 развился из общего предка с CoV крупного рогатого скота (КРС) при преодолении межвидового барьера в 1890-х гг. (Vijgen et al., 2005, 2006).
На сегодня одна из наиболее обсуждаемых тем – возможность участия животных-компаньонов в цепи трансмиссии эмерджентных коронавирусных инфекций, поскольку эти животные находятся в самом близком контакте с человеком. Пока не существует достаточной доказательной базы для утверждения, что мелкие домашние животные задействованы в возникновении и распространении эмерджентных CoV.
Единственный достоверно известный CoV, который вызывает инфекционно-воспалительное заболевание у домашних кошек Felis silvestris catus – кошачий CoV, который относится к роду Alphacoronavirus и подразделяется на два типа на основании генетического и антигенного различий белка-“шипа” (spike protein) (Jaimes et al., 2018; Felten, Hartmann, 2019). В реальности большинство коронавирусных перитонитов у кошек вызвано кошачьим CoV I типа, в то время как II тип относительно менее распространен, поскольку установлено, что этот CoV образовался при генетической рекомбинации с собачьим CoV, что делает его менее контагиозным для кошек (Le Poder, 2011). Это подтверждается недавним исследованием Жао с соавт., в котором при серологическом скрининге было обнаружено, что 75 (54.7%) и 26 (19.9%) из 137 образцов плазмы были серопозитивны по отношению к I и II типам кошачьего CoV соответственно. При этом все образцы плазмы кошек, серопозитивные по отношению к II типу, были также позитивны по отношению к I типу, что указывает на феномен перекрестной реактивности, поскольку не все животные из этой группы были инфицированы кошачьим CoV I типа (Zhao et al., 2019).
Кошек иногда используют в качестве модельных экспериментальных животных для воспроизведения различных вирусных инфекций в целях изучения их патологических аспектов, возможности трансмиссии между особями и апробации методов лечения. Так, в 1984–1985 гг. были описаны успешные методики экспериментального воспроизведения коронавирусной инфекции кошек с помощью собачьего CoV и человеческого HCoV-229E (Barlough et al., 1984, 1985). В 2003 г. были опубликовали экспериментальные данные, согласно которым у кошек через 4 и 6 сут после интратрахеальной инокуляции 106 ед. 50%-ной инфицирующей дозы SARS-CoV в образцах назальных мазков был обнаружен этот вирус в отсутствие типичной симптоматики. Это свидетельствует о подверженности организма этих животных заражению еще одним человеческим CoV. Были описаны также случаи трансмиссии SARS-CoV другим лабораторным кошкам при помещении их в закрытую клетку с зараженной особью (Martina et al., 2003). При проведении аналогичного экспериментального исследования были подтверждены эти данные и дополнительно описаны альтерации дыхательных путей при воспроизведении инфицирования SARS-CoV, характерные для трахеобронхоаденита (van den Brand et al., 2008).
Отметим, что все модельные работы по воспроизведению коронавирусных инфекций на кошках сопровождаются сравнением с контрольной и экспериментальной группами хорьков Mustela putorius, которые также довольно популярные животные-компаньоны. У этих животных во всех работах отмечаются более выраженные воспалительные реакции и иммунные ответы, которые по характеру ближе к таковым у человека, что делает хорьков более подходящей моделью для изучения человеческой патологии (Martina et al., 2003; van den Brand et al., 2008; Gong et al., 2018). Однако при попытке воспроизвести коронавирусную инфекцию с помощью MERS-CoV хорьки оказывались интактными к вирусному агенту (Raj et al., 2014b). Было обнаружено, что другие лабораторные животные, а именно новозеландские белые кролики Oryctolagus cuniculus, могут подходить для in vitro моделирования респираторной инфекции MERS-CoV (Haagmans et al., 2015), что может снизить необходимость применения генетически модифицированных мышей Mus musculus для этих целей (Agrawal et al., 2015).
Сразу после вспышки новой коронавирусной инфекции в декабре 2019 г. в г. Ухань китайской провицнции Хубэй начались исследования по идентификации вирусного агента, которого позже назвали SARS-CoV-2 из-за генетических и фенотипических сходств с SARS-CoV, а также ввиду схожести клинического течения вызываемых ими заболеваний (Gorbalenya et al., 2020; Yang et al., 2020). При обнаружении того, что инвазия клетки SARS-CoV-2 происходит посредством взаимодействия с ангиотензинпревращающим ферментом-2 (АПФ2), ученые выдвинули гипотезу, что этот вирус можно изучать на тех же модельных животных, которые использовались для моделирования инфекции, вызванной SARS-CoV. Ши с соавт. провели обширное экспериментальное исследование, в которое включили множество видов мелких домашних и сельскохозяйственных животных, которым интраназально инокулировали 105 бляшкообразующих единиц SARS-CoV-2. По аналогии с SARS-CoV только кошки (в меньшей степени) и хорьки (в большей степени) реагировали на SARS-CoV-2 (Shi et al., 2020), что, несомненно, связано со сходством строения АПФ2 этих животных и человека (Guo et al., 2008). Результаты исследования Ши с соавт. вызвали множество вопросов о возможности участия мелких домашних животных, в частности кошек, в путях трансмиссии SARS-CoV-2.
Мы считаем нужным указать на неестественность процесса заражения SARS-CoV-2 кошек в этом экспериментальном исследовании, так как вероятность возникновения реальных условий, при которых напрямую интраназально будет введена экстремально большая доза вирусного патогена, крайне низка. В исследовании получены ценные данные о возможности использования кошек и хорьков в качестве модельных животных, но не более. Для сравнения считаем нужным привести ранее упомянутое исследование Хаагманс с соавт., в котором кроликам также интраназально и интратрахеально вводили 1 × 106 и 4 × 106 ед. 50%‑ной инфицирующей дозы MERS-CoV соответственно (Haagmans et al., 2015). Популяция кроликов во всем мире неоспоримо больше популяции одногорбых верблюдов (DiVincenti et al., 2016; Sazmand et al., 2019), и за 8 лет с момента идентификации у первого больного MERS-CoV ни одного случая трансмиссии этого вируса между кроликами и людьми не задокументировано. Поэтому экстраполировать экспериментальные данные, полученные в лабораторных условиях, на реальность нецелесообразно. Реагирование любого животного на человеческий вирусный патоген при исключении участия этого животного в трансмиссионной цепи свидетельствует лишь о схожести молекулярно-биохимических процессов в двух организмах. Это также позволяет использовать животное в качестве потенциального модельного объекта для изучения способов воздействия на вирусную инфекцию человека.
Однако с кошками ситуация намного усложняется ввиду публикаций результатов массовых серологических исследований, одно из которых было проведено в г. Ухань. Было обнаружено, что 15 из 102 образцов плазмы кошек, взятых во время вспышки COVID-19, были серопозитивны по отношению к домену связывания рецептора SARS-CoV-2, что указывает на их возможное инфицирование, в то время как 39 образцов, отобранных между мартом и маем 2019 г. были серонегативны. Важно то, что наибольший титр нейтрализации был отмечен у трех кошек, хозяева которых болели COVID-19 (Zhang et al., 2020). В это же время были опубликованы результаты исследования, согласно которым у всех 9 включенных в исследования кошек не было обнаружено ни признаков инфицирования SARS-CoV с помощью ОТ-ПЦР, ни антител против этого вируса в их крови на основе иммунопреципитации (Temmam et al., 2020). Также не стоит оставлять без внимания отмеченные в СМИ случаи заражения в зоопарках разных представителей семейства кошачьих (Felidae) SARS-CoV-2, хотя сведения об использованных методах исследования были ограничены. При обсуждении вышеописанных исследований необходимо руководствоваться принципами медицины и ветеринарии, основанными на доказательствах, а значит не делать окончательных выводов о возможности заражения кошек SARS-CoV-2 от человека в реальных условиях и тем более об обратной трансмиссии.
Тем не менее мы допускаем возможность инфицирования представителей семейства кошачьих новым CoV ввиду сходства молекулярного строения рецепторной мишени этого вируса, но только в условиях очень тесного и повторяющегося контакта с зараженным человеком, когда суммарный титр вируса, передающегося воздушно-капельным путем, достигает значений близких к полученным при экспериментальном моделировании. Однако в реальных условиях шанс возникновения подобного стечения обстоятельств маловероятен. Также нельзя забывать о первой вспышке коронавирусной инфекции, вызванной SARS-CoV, к которому кошки при экспериментальном инфицировании также оказались восприимчивы (Martina et al., 2003). За 18 лет в научной литературе не описано ни одного подтвержденного случая распространения SARS-CoV кошками среди региональных популяций и тем более случаев обратной трансмиссии этой инфекции человеку.
Мы убеждены, что если инфицирование SARS-CoV-2 кошек подтвердится, то эти животные будут для этого вируса биологическим тупиком. Также мы считаем, что для обсуждения факта носительства SARS-CoV-2 и его трансмиссии кошками критически необходимо проведение полномасштабного когортного клинического исследования, притом слепого и рандомизированного, что исключит влияние человеческого фактора на результаты, поскольку неверная их интерпретация может привести к необратимым последствиям. Кроме того, в этом исследовании необходимо применить методы обнаружения субгеномной РНК вируса, что с наибольшей вероятностью укажет на факт репликации SARS-CoV-2 в клетках животных.
Намного интереснее ситуация с собаками Canis lupus familiaris. На настоящий момент установлены два CoV собак: один из них относится к роду Alphacoronavirus и вызывает воспалительные кишечные заболевания (Decaro, Buonavoglia., 2008, 2011), другой – к роду Betacoronavirus и ответствен за поражение респираторных органов (Erles et al., 2007; Lu et al., 2017). Похоже, что у обоих вирусов наблюдаются высокая репликационная активность и закономерное стремительное эволюционное развитие. Собачий кишечный CoV обнаружили в 1971 г. Бинн с соавт., которые выделили его у служебных собак с острым вирусным энтеритом в Германии (Binn et al., 1974). Позже на основании генотипических различий он был разделен на два типа (Decaro et al., 2010a). Важно отметить, что типы собачьего кишечного CoV гомологичны аналогичным типам кошачьего CoV (Decaro et al., 2010b), а II тип в свою очередь подразделен на два подтипа IIа и IIb на основании генетических различий, возникших вследствие рекомбинации с вирусом трансмиссивного гастроэнтерита свиней Sus scrofa domesticus и приобретения общих качеств между ними (Decaro et al., 2009). Собачий респираторный CoV обнаружили в 2003 г. в Великобритании. При определении эволюционного происхождения этого вируса было установлено близкое сродство собачьего респираторного CoV c CoV КРС и HCoV-OC43, что также свидетельствует об одном общем вирусе-предшественнике (Erles, Brownlie, 2008). Удалось экспериментально воспроизвести инфекцию у щенков с помощью CoV КРС, на основании чего ученые предположили, что предшественник собачьего респираторного вируса мог перейти к собакам от КРС (Kaneshima et al., 2007), что перекликается с предположением об аналогичном происхождении HCoV-OC43 (Vijgen et al., 2005, 2006).
Таким образом, на основании данных о гомологичности между CoV разных видов можно утверждать, что собаки участвуют в разных путях межвидовой трансмиссии этих вирусов. И точно так же, как с кошками, научное сообщество задалось вопросом о возможности заражения SARS-CoV-2 собак и участии их в переносе инфекции. На наш взгляд, наиболее интересна публикация Сиа, который выдвинул гипотезу о том, что бродячие собаки провинции Хубэй могут быть естественными резервуарами предшественника SARS-CoV-2 и что высокое содержание противовирусного белка ZAP в кишечной ткани собак могло спровоцировать развитие потенциального вируса-предшественника в сторону приобретения способности сопротивляться ZAP-ассоциированному иммунному ответу посредством адаптационного уменьшения CpG-динуклеотидов в вирусной РНК (Xia, 2020).
Гипотеза представляет определенный интерес, и мы уверены, что принцип подобного взаимоотношения между белками ZAP хозяина и CpG-динуклеотидами CoV может найти свое место в биоинженерии. Однако мы убеждены, что реальность этой гипотезы происхождения SARS-CoV-2 маловероятна по ряду причин:
во-первых, во время обсуждения автор не берет во внимание исследование, в котором собаки оказались невосприимчивыми к SARS-CoV-2 при интраназальной инокуляции экстремально высокого количества бляшкообразующих единиц вируса (Shi et al., 2020), в то время как у животных, которые предполагались в качестве резервуаров CoV, при экспериментальном моделировании наблюдались явно выраженные признаки инфицирования – циветы оказались восприимчивы к SARS-CoV (Xiao et al., 2008), а одногорбые верблюды – к MERS-CoV (Adney et al., 2014);
во-вторых, за несколько месяцев пандемии у собак провинции Хубэй не идентифицировали SARS-подобных CoV и не обнаружили антител к этим вирусам. Ввиду упомянутых выше фактов мы убеждены, что собаки не задействованы в трансмиссии SARS-CoV-2 и даже возможность их носительства в качестве биологического тупика крайне маловероятна.
Необходимо отметить, что пять из семи первичных резервуаров зоонозных CoV, в том числе наиболее опасных SARS-CoV, MERS-CoV и SARS-CoV-2, – различные представители отряда рукокрылых. Однако это не означает, что только эти животные представляют эпидемиологическую опасность для человека: грызуны (Rodentia), как и другие отряды млекопитающих, также могут быть потенциальным резервуаром для новой вспышки коронавирусной инфекции.
При анализе эволюционных путей развития человеческих CoV нами было отмечено несколько закономерностей, на основании чего предложены два сценария возникновения эмерджентных CoV, различающихся степенью человеческого вмешательства в дикую природу (рис. 1). Первый сценарий не подразумевает выраженного влияния людей на цепь трансмиссий CoV, а именно преднамеренное вторжение в жизнедеятельность диких животных, приводящее к встрече видов животных, ареалы которых в естественных условиях не пересекаются. Так, достоверно известно, что MERS-CoV циркулировал в популяции одногорбых верблюдов в течение десятилетий, что подтверждается меньшим генетическим сродством с потенциальными предшественниками CoV летучих мышей (75–87%) по сравнению с SARS-CoV (95%) (Mohd et al., 2016; Lau et al., 2018; Luo et al., 2018). На устойчивое развитие MERS-CoV в организме верблюдов указывают также выявление у животных и человека идентичных на 100% вирусов, а также необходимость контакта животного-резервуара для заражения человека и самая высокая человеческая летальность (34.4%) (Petrosillo et al., 2020). Все это свидетельствует о закономерной и стабильной эволюции MERS-CoV: вирус недавно попал в организм человека, что объясняет его низкую адаптацию к новому хозяину, выражающуюся в сравнительно малой способности к трансмиссии между людьми (Petrosillo et al., 2020).
Рис. 1.
Схематическое изображение двух сценариев возникновения эмерджентных коронавирусных инфекций. I – cценарий развития MERS-CoV; II – сценарий развития SARS-CoV и, вероятно, SARS-CoV-2.
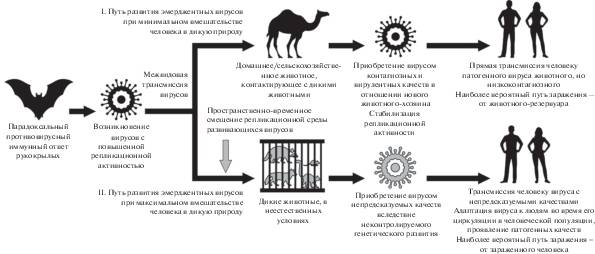
Мы предполагаем, что развитие SARS-CoV по сравнению с развитием MERS-CoV произошло намного стремительнее из-за вмешательства человека в дикую природу. Известно, что один из наиболее вероятных очагов первичной инфекции – рынок животных Шэньчжэн, условия которого идеальны для смешения микробиоты различных видов. Клеточное содержание на малой площади большого количества животных, изъятых из дикой природы из отдаленных ареалов, неблагоприятно сказывается на резистентности их иммунной системы из-за хронического стресса, что на фоне несоблюдения санитарных условий способствует межвидовой трансмиссии вирусной микрофлоры и повышенной репликационной активности CoV при адаптации к новым хозяевам.
Подобная сублимация – пространственно-временнóе смещение из-за встречи вирусной микробиоты животных, которые без вмешательства человека в природную среду могли никогда не контактировать между собой. Все вышеизложенное делает невозможным предугадать вектор генетического развития CoV в таких условиях, что и представляет опасность возникновения CoV с неизвестными качествами. Кроме того, в результате такой бесконтрольной трансмиссии CoV не успевает адаптироваться к какому-либо одному животному, что не снижает его способность к преодолению межвидового барьера и не стабилизирует репликационную активность, приобретенную благодаря рукокрылым. Это предоставляет возможность CoV с неизвестными патогенными свойствами быстро адаптироваться к организму нового хозяина в лице человека и приобрести устойчивую внутривидовую контагиозность. Примеры этого сценария – SARS-CoV и, вероятно, SARS-CoV-2, поскольку они более близки к потенциальным вирусам-предшественникам рукокрылых (95 и 96.3%), их вероятные промежуточные хозяева содержались на китайских рынках животных (циветы, енотовидные собаки – SARS-CoV; яванский панголин Manis javanica – SARS-CoV-2), а также их контагиозность между людьми заметно выше, чем у MERS-CoV, а летальность ниже (9.5 и 2.3% соответственно) (Lam et al., 2020; Yang et al., 2020).
Самой эффективной мерой предотвращения будущих вспышек, на наш взгляд, является биоветеринарный и санитарный контроль рынков животных, причем не только в Китае. После исключения условий, которые создают благоприятный фон для непредсказуемой эволюции патогенных CoV, у нас появится больше ресурсов для исследования стабильных трансмиссионных цепей, таких как MERS-CoV, и методов их контроля.
Другой потенциальный и результативный способ профилактики распространения коронавирусных инфекций – пробиотические бактерии ввиду их способности подавлять вирусную репликационную активность, а также оказывать противовирусное влияние с помощью продуцируемых ими молочной кислоты, перекиси водорода и бактериоцинов (Sekar et al., 2016; Kanmani et al., 2018; Abdelhamid et al., 2019). Кроме того, при исследовании микробиоты кошек было обнаружено, что бактерии рода Lactobacillus могут оказать антагонистическое влияние на кошачий кишечный CoV (Mingmongkolchai, Panbangred., 2018), что указывает на необходимость проведения полномасштабных исследований возможности модулирования здоровья животных с помощью пробиотиков. Применение пробиотиков в сельском хозяйстве позволит предотвратить возникновение эмерджентных коронавирусных инфекций от КРС и других животных, которые могут оказаться резервуаром инфекции, что также снизит вероятность повторения первого варианта предложенного нами сценария.
ЗАКЛЮЧЕНИЕ
Коронавирусы животных представляют эпидемиологическую опасность ввиду их повышенной способности к межвидовой трансмиссии, репликационной и рекомбинаторной активности. У множества видов животных обнаруживаются гомологичные коронавирусы с общими качествами, что указывает на их непрерывное эволюционное развитие. Однако наибольший вклад в создание условий для возникновения новых эмерджентных коронавирусов вносит человек посредством вторжения в природу, изъятия диких животных из их ареала, обеспечения контактов между видами, которые в естественных условиях никогда не встречаются, и закономерного создания новых трансмиссионных путей коронавирусов между дикими животными и человеком.
В настоящий момент не существует доказательной базы того, что животные-компаньоны могут быть переносчиками новой коронавирусной инфекции. Однако не исключена возможность того, что представители семейства кошачьих могут оказаться биологическим тупиком в глобальных путях трансмиссии SARS-CoV-2. Эта тема требует полномасштабных когортных клинических исследований.
Подобно сингулярному состоянию перед Большим взрывом, человек, вмешиваясь в дикую природу, сталкивает в одной пространственно-временнóй точке разные виды животных и их вирусы, что провоцирует эмерджентность биосферы с непредсказуемыми последствиями.
Работа выполнена при финансовой поддержке Правительства Российской Федерации (соглашение № 075-15-2019-1880).
Список литературы
Abdelhamid A.G., El-Masry S.S., El-Dougdoug N.K. Probiotic Lactobacillus and Bifidobacterium strains possess safety characteristics, antiviral activities and host adherence factors revealed by genome mining // EPMA J. 2019. V. 10. № 4. P. 337–350. https://doi.org/10.1007/s13167-019-00184-z
Adney D.R., van Doremalen N., Brown V.R., Bushmaker T., Scott D., de Wit E., Bowen R.A., Munster V.J. Replication and shedding of MERS-CoV in upper respiratory tract of inoculated dromedary camels // Emerg. Infect. Dis. 2014. V. 20. № 12. P. 1999–2005. https://doi.org/10.3201/eid2012.141280
Agrawal A.S., Garron T., Tao X., Peng B.H., Wakamiya M., Chan T.S., Couch R.B., Tseng C.T. Generation of a transgenic mouse model of Middle East respiratory syndrome coronavirus infection and disease // J. Virol. 2015. V. 89. № 7. P. 3659–3670. https://dx.doi.org/10.1128%2FJVI.03427-14
Ahn M., Anderson D.E., Zhang Q., Tan C.W., Lim B.L., Luko K., Wen M., Chia W.N., Mani S., Wang L.C., Ng J.H.J., Sobota R.M., Dutertre C.A., Ginhoux F., Shi Z.L., Irving A.T., Wang L.F. Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host // Nat. Microbiol. 2019. V. 4. № 5. P. 789–799. https://dx.doi.org/10.1038%2Fs41564-019-0371-3
Barlough J.E., Johnson-Lussenburg C.M., Stoddart C.A., Jacobson R.H., Scott F.W. Experimental inoculation of cats with human coronavirus 229E and subsequent challenge with feline infectious peritonitis virus // Can. J. Comp. Med. 1985. V. 49. № 3. P. 303–307.
Barlough J.E., Stoddart C.A., Sorresso G.P., Jacobson R.H., Scott F.W. Experimental inoculation of cats with canine coronavirus and subsequent challenge with feline infectious peritonitis virus // Lab. Anim. Sci. 1984. V. 34. № 6. P. 592–597.
Binn L.N., Lazar E.C., Keenan K.P., Huxsoll D.L., Marchwicki R.H., Strano A.J. Recovery and characterization of a coronavirus from military dogs with diarrhea // Proc. Annu. Meet. US Anim. Health. Assoc. 1974. V. 78. P. 359–366.
Brook C.E., Boots M., Chandran K., Dobson A.P., Drosten C., Graham A.L., Grenfell B.T., Müller M.A., Ng M., Wang L.F., van Leeuwen A. Accelerated viral dynamics in bat cell lines, with implications for zoonotic emergence // Elife. 2020. V. 9. e48401. https://dx.doi.org/10.7554%2FeLife.48401
Carman P.S., Hazlett M.J. Bovine coronavirus infection in Ontario 1990-1991 // Can. Vet. J. 1992. V. 33. № 12. P. 812–814.
Chu H., Chan J.F.W., Yuen T.T.T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.C.Y., Tsang J.O.L., Huang X., Chai Y., Yang D., Hou Y., Chik K.K.H., Zhang X., Fung A.Y.F., Tsoi H.W., Cai J.P., Chan W.M., Ip J.D., Chu A.W.H., Zhou J., Lung D.C., Kok K.H., To K.K.W., Tsang O.T.Y., Chan K.H., Yuen K.Y. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study // The Lancet Microbe. 2020. V. 1. № 1. P. e14–e23. https://dx.doi.org/10.1016%2FS2666-5247(20)30004-5
Collins S.E., Mossman K.L. Danger, diversity and priming in innate antiviral immunity // Cytokine Growth Factor Rev. 2014. V. 25. № 5. P. 525–531. https://doi.org/10.1016/j.cytogfr.2014.07.002
Corman V.M., Muth D., Niemeyer D., Drosten C. Hosts and Sources of Endemic Human Coronaviruses // Adv. Virus. Res. 2018. V. 100. P. 163–188. https://dx.doi.org/10.1016%2Fbs.aivir.2018.01.001
Corman V.M., Baldwin H.J., Tateno A.F., Zerbinati R.M., Annan A., Owusu M., Nkrumah E.E., Maganga G.D., Oppong S., Adu-Sarkodie Y., Vallo P., da Silva Filho L.V., Leroy E.M., Thiel V., van der Hoek L., Poon L.L., Tschapka M., Drosten C., Drexler J.F. Evidence for an Ancestral Association of Human Coronavirus 229E with Bats // J. Virol. 2015. V. 89. № 23. P. 11858–11870. https://dx.doi.org/10.1128%2FJVI.01755-15
Crossley B.M., Mock R.E., Callison S.A., Hietala S.K. Identification and characterization of a novel alpaca respiratory coronavirus most closely related to the human coronavirus 229E // Viruses. 2012. V. 4. № 12. P. 3689–3700. https://dx.doi.org/10.3390%2Fv4123689
Crossley B.M., Barr B.C., Magdesian K.G., Ing M., Mora D., Jensen D., Loretti A.P., McConnell T., Mock R. Identification of a novel coronavirus possibly associated with acute respiratory syndrome in alpacas (Vicugna pacos) in California, 2007 // J. Vet. Diagn. Invest. 2010. V. 22. № 1. P. 94–97. https://doi.org/10.1177/104063871002200118
Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses // Nat. Rev. Microbiol. 2019. V. 17. № 3. P. 181–192. https://dx.doi.org/10.1038%2Fs41579-018-0118-9
Decaro N., Buonavoglia C. An update on canine coronaviruses: viral evolution and pathobiology // Vet. Microbiol. 2008. V. 132. № 3–4. P. 221–234. https://dx.doi.org/10.1016%2Fj.vetmic.2008.06.007
Decaro N., Buonavoglia C. Canine coronavirus: not only an enteric pathogen // Vet. Clin. North Am. Small Anim. Pract. 2011. V. 41. № 6. P. 1121–1132. https://doi.org/10.1016/j.cvsm.2011.07.005
Decaro N., Mari V., Elia G., Addie D.D., Camero M., Lucente M.S., Martella V., Buonavoglia C. Recombinant canine coronaviruses in dogs, Europe // Emerg. Infect. Dis. 2010a. V. 16. № 1. P. 41–47. https://dx.doi.org/10.3201%2Feid1601.090726
Decaro N., Mari V., Campolo M., Lorusso A., Camero M., Elia G., Martella V., Cordioli P., Enjuanes L., Buonavoglia C. Recombinant canine coronaviruses related to transmissible gastroenteritis virus of Swine are circulating in dogs // J. Virol. 2009. V. 83. № 3. P. 1532–1537. https://dx.doi.org/10.1128%2FJVI.01937-08
Decaro N., Elia G., Martella V., Campolo M., Mari V., Desario C., Lucente M.S., Lorusso E., Kanellos T., Gibbons R.H., Buonavoglia C. Immunity after natural exposure to enteric canine coronavirus does not provide complete protection against infection with the new pantropic CB/05 strain // Vaccine. 2010b. V. 28. № 3. P. 724–729. https://dx.doi.org/10.1016%2Fj.vaccine.2009.10.077
Decaro N., Campolo M., Desario C., Cirone F., D’Abramo M., Lorusso E., Greco G., Mari V., Colaianni M.L., Elia G., Martella V., Buonavoglia C. Respiratory disease associated with bovine coronavirus infection in cattle herds in Southern Italy // J. Vet. Diagn. Invest. 2008. V. 20. № 1. P. 28–32. https://doi.org/10.1177/104063870802000105
DiVincenti L., Jr., Rehrig A.N. The Social Nature of European Rabbits (Oryctolagus cuniculus) // J. Am. Assoc. Lab. Anim. Sci. 2016. V. 55. № 6. P. 729–736.
Drexler J.F., Corman V.M., Müller M.A., Maganga G.D., Vallo P., Binger T., Gloza-Rausch F., Cottontail V.M., Rasche A., Yordanov S., Seebens A., Knörnschild M., Oppong S., Adu Sarkodie Y., Pongombo C., Lukashev A.N., Schmidt-Chanasit J., Stöcker A., Carneiro A.J., Erbar S., Maisner A., Fronhoffs F., Buettner R., Kalko E.K., Kruppa T., Franke C.R., Kallies R., Yandoko E.R., Herrler G., Reusken C., Hassanin A., Krüger D.H., Matthee S., Ulrich R.G., Leroy E.M., Drosten C. Bats host major mammalian paramyxoviruses // Nat. Commun. 2012. V. 3. P. 796. https://dx.doi.org/10.1038%2Fncomms1796
Erles K., Brownlie J. Canine respiratory coronavirus: an emerging pathogen in the canine infectious respiratory disease complex // Vet. Clin. North Am. Small Anim. Pract. 2008. V. 38. № 4. P. 815–825. https://dx.doi.org/10.1016%2Fj.cvsm.2008.02.008
Erles K., Shiu K.B., Brownlie J. Isolation and sequence analysis of canine respiratory coronavirus // Virus. Res. 2007. V. 124. № 1–2. P. 78–87. https://dx.doi.org/10.1016%2Fj.virusres.2006.10.004
Felten S., Hartmann K. Diagnosis of Feline Infectious Peritonitis: A Review of the Current Literature // Viruses. 2019. V. 11. № 11. P. 1068. https://dx.doi.org/10.3390%2Fv11111068
Forbes K.M., Webala P.W., Jääskeläinen A.J., Abdurahman S., Ogola J., Masika M.M., Kivistö I., Alburkat H., Plyusnin I., Levanov L., Korhonen E.M., Huhtamo E., Mwaengo D., Smura T., Mirazimi A., Anzala O., Vapalahti O., Sironen T. Bombali Virus in Mops condylurus Bat, Kenya // Emerg. Infect. Dis. 2019. V. 25. № 5. P. 955–957. https://dx.doi.org/10.3201%2Feid2505.181666
Goldstein T., Anthony S.J., Gbakima A., Bird B.H., Bangura J., Tremeau-Bravard A., Belaganahalli M.N., Wells H.L., Dhanota J.K., Liang E., Grodus M., Jangra R.K., DeJesus V.A., Lasso G., Smith B.R., Jambai A., Kamara B.O., Kamara S., Bangura W., Monagin C., Shapira S., Johnson C.K., Saylors K., Rubin E.M., Chandran K., Lipkin W.I., Mazet J.A.K. The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses // Nat. Microbiol. 2018. V. 3. № 10. P. 1084–1089. https://dx.doi.org/10.1038%2Fs41564-018-0227-2
Gong S.R., Bao L.L. The battle against SARS and MERS coronaviruses: Reservoirs and Animal Models // Anim. Model Exp. Med. 2018. V. 1. № 2. P. 125–133. https://dx.doi.org/10.1002%2Fame2.12017
Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2 // Nat. Microbiol. 2020. V. 5. № 4. P. 536–544. https://doi.org/10.1038/s41564-020-0695-z
Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China // Science. 2003. V. 302. № 5643. P. 276–278. https://doi.org/10.1126/science.1087139
Guo H., Guo A., Wang C., Yan B., Lu H., Chen H. Expression of feline angiotensin converting enzyme 2 and its interaction with SARS-CoV S1 protein // Res. Vet. Sci. 2008. V. 84. № 3. P. 494–496. https://dx.doi.org/10.1016%2Fj.rvsc.2007.05.011
Haagmans B.L., van den Brand J.M., Provacia L.B., Raj V.S., Stittelaar K.J., Getu S., de Waal L., Bestebroer T.M., van Amerongen G., Verjans G.M., Fouchier R.A., Smits S.L., Kuiken T., Osterhaus A.D. Asymptomatic Middle East respiratory syndrome coronavirus infection in rabbits // J. Virol. 2015. V. 89. № 11. P. 6131–6135. https://dx.doi.org/10.1128%2FJVI.00661-15
Hawkins J.A., Kaczmarek M.E., Müller M.A., Drosten C., Press W.H., Sawyer S.L. A metaanalysis of bat phylogenetics and positive selection based on genomes and transcriptomes from 18 species // Proc. Natl Acad. Sci. USA. 2019. V. 116. № 23. P. 11351–11360. https://dx.doi.org/10.1073%2Fpnas.1814995116
Heckert R.A., Saif L.J., Hoblet K.H., Agnes A.G. A longitudinal study of bovine coronavirus enteric and respiratory infections in dairy calves in two herds in Ohio // Vet. Microbiol. 1990. V. 22. № 2–3. P. 187–201. https://dx.doi.org/10.1016%2F0378-1135(90)90106-6
Jaimes J.A., Whittaker G.R. Feline coronavirus: Insights into viral pathogenesis based on the spike protein structure and function // Virology. 2018. V. 517. P. 108–121. https://dx.doi.org/10.1016%2Fj.virol.2017.12.027
Ji W., Wang W., Zhao X., Zai J., Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV // J. Med. Virol. 2020. V. 92. № 4. P. 433–440. https://dx.doi.org/10.1002%2Fjmv.25682
Kaneshima T., Hohdatsu T., Hagino R., Hosoya S., Nojiri Y., Murata M., Takano T., Tanabe M., Tsunemitsu H., Koyama H. The infectivity and pathogenicity of a group 2 bovine coronavirus in pups // J. Vet. Med. Sci. 2007. V. 69. № 3. P. 301–303. https://doi.org/10.1292/jvms.69.301
Kanmani P., Albarracin L., Kobayashi H., Hebert E.M., Saavedra L., Komatsu R., Gatica B., Miyazaki A., Ikeda-Ohtsubo W., Suda Y., Aso H., Egusa S., Mishima T., Salas-Burgos A., Takahashi H., Villena J., Kitazawa H. Genomic characterization of Lactobacillus delbrueckii TUA4408L and evaluation of the antiviral activities of its extracellular polysaccharides in porcine intestinal epithelial cells // Front. Immunol. 2018. V. 9. P. 2178. https://doi.org/10.3389/fimmu.2018.02178
Lam T.T., Jia N., Zhang Y.W., Shum M.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., Li W.J., Jiang B.G., Wei W., Yuan T.T., Zheng K., Cui X.M., Li J., Pei G.Q., Qiang X., Cheung W.Y., Li L.F., Sun F.F., Qin S., Huang J.C., Leung G.M., Holmes E.C., Hu Y.L., Guan Y., Cao W.C. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins // Nature. 2020. V. 583. № 7815. P. 282–285. https://doi.org/10.1038/s41586-020-2169-0
Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats // Proc. Natl Acad. Sci. USA. 2005. V. 102. № 39. P. 14040–14045. https://dx.doi.org/10.1073%2Fpnas.0506735102
Lau S.K.P., Zhang L., Luk H.K.H., Xiong L., Peng X., Li K.S.M., He X., Zhao P.S., Fan R.Y.Y., Wong A.C.P., Ahmed S.S., Cai J.P., Chan J.F.W., Sun Y., Jin D., Chen H., Lau T.C.K., Kok R.K.H., Li W., Yuen K.Y., Woo P.C.Y. Receptor usage of a novel bat lineage C betacoronavirus reveals evolution of Middle East respiratory syndrome-Related coronavirus spike proteins for human dipeptidyl peptidase 4 binding // J. Infect. Dis. 2018. V. 218. № 2. P. 197–207. https://dx.doi.org/10.1093%2Finfdis%2Fjiy018
Le Poder S. Feline and canine coronaviruses: common genetic and pathobiological features // Adv. Virol. 2011. V. 2011. P. 609465. https://dx.doi.org/10.1155%2F2011%2F609465
Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses // Science. 2005. V. 310. № 5748. P. 676–679. https://doi.org/10.1126/science.1118391
Li X., Luk H.K.H., Lau S.K.P., Woo P.C.Y. Human Coronaviruses: General Features // Ref. Mod. Biomed. Sci. 2019. P. B978-0-12-801238-3.95704-0. https://dx.doi.org/10.1016%2FB978-0-12-801238-3.95704-0
Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding // Lancet. 2020. V. 395. № 10224. P. 565–574. https://dx.doi.org/10.1016%2FS0140-6736(20)30251-8
Lu S., Wang Y., Chen Y., Wu B., Qin K., Zhao J., Lou Y., Tan W. Discovery of a novel canine respiratory coronavirus support genetic recombination among betacoronavirus1 // Virus. Res. 2017. V. 237. P. 7–13. https://dx.doi.org/10.1016%2Fj.virusres.2017.05.006
Luo C.M., Wang N., Yang X.L., Liu H.Z., Zhang W., Li B., Hu B., Peng C., Geng Q.B., Zhu G.J., Li F., Shi Z.L. Discovery of novel bat coronaviruses in South China that use the same receptor as Middle East respiratory syndrome coronavirus // J. Virol. 2018. V. 92. № 13. P. e00116–00118. https://dx.doi.org/10.1128%2FJVI.00116-18
Martina B.E., Haagmans B.L., Kuiken T., Fouchier R.A., Rimmelzwaan G.F., Van Amerongen G., Peiris J.S., Lim W., Osterhaus A.D. Virology: SARS virus infection of cats and ferrets // Nature. 2003. V. 425. № 6961. P. 915. https://dx.doi.org/10.1038%2F425915a
Milewska A., Nowak P., Owczarek K., Szczepanski A., Zarebski M., Hoang A., Berniak K., Wojarski J., Zeglen S., Baster Z., Rajfur Z., Pyrc K. Entry of Human Coronavirus NL63 into the cell // J. Virol. 2018. V. 92. № 3. P. e01933–01917. https://dx.doi.org/10.1128%2FJVI.01933-17
Milne-Price S., Miazgowicz K.L., Munster V.J. The emergence of the Middle East respiratory syndrome coronavirus // Pathog. Dis. 2014. V. 71. № 2. P. 121–136. https://dx.doi.org/10.1111%2F2049-632X.12166
Mingmongkolchai S., Panbangred W. Bacillus probiotics: an alternative to antibiotics for livestock production // J. Appl. Microbiol. 2018. V. 124. № 6. P. 1334–1346. https://doi.org/10.1111/jam.13690
Mohd H.A., Al-Tawfiq J.A., Memish Z.A. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) origin and animal reservoir // Virol. J. 2016. V. 13. P. 87. https://dx.doi.org/10.1186%2Fs12985-016-0544-0
Ogando N.S., Ferron F., Decroly E., Canard B., Posthuma C.C., Snijder E.J. The Curious Case of the Nidovirus Exoribonuclease: Its Role in RNA Synthesis and Replication Fidelity // Front. Microbiol. 2019. V. 10. P. 1813. https://dx.doi.org/10.3389%2Ffmicb.2019.01813
Omrani A.S., Al-Tawfiq J.A., Memish Z.A. Middle East respiratory syndrome coronavirus (MERS-CoV): animal to human interaction // Pathog. Glob. Health. 2015. V. 109. № 8. P. 354–362. https://dx.doi.org/10.1080%2F20477724.2015.1122852
Pavlovich S.S., Lovett S.P., Koroleva G., Guito J.C., Arnold C.E., Nagle E.R., Kulcsar K., Lee A., Thibaud-Nissen F., Hume A.J., Mühlberger E., Uebelhoer L.S., Towner J.S., Rabadan R., Sanchez-Lockhart M., Kepler T.B., Palacios G. The Egyptian Rousette Genome Reveals Unexpected Features of Bat Antiviral Immunity // Cell. 2018. V. 173. № 5. P. 1098–1110.e18. https://dx.doi.org/10.1016%2Fj.cell.2018.03.070
Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis // Nat. Rev. Microbiol. 2009. V. 7. P. 439–450. https://doi.org/10.1038/nrmicro2147
Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? // Clin. Microbiol. Infect. 2020. V. 26. № 6. P. 729–734. https://dx.doi.org/10.1016%2Fj.cmi.2020.03.026
Raj V.S., Smits S.L., Provacia L.B., van den Brand J.M., Wiersma L., Ouwendijk W.J., Bestebroer T.M., Spronken M.I., van Amerongen G., Rottier P.J., Fouchier R.A., Bosch B.J., Osterhaus A.D., Haagmans B.L. Adenosine deaminase acts as a natural antagonist for dipeptidyl peptidase 4-mediated entry of the Middle East respiratory syndrome coronavirus // J. Virol. 2014a. V. 88. № 3. P. 1834–1838. https://dx.doi.org/10.1128%2FJVI.02935-13
Raj V.S., Farag E.A., Reusken C.B., Lamers M.M., Pas S.D., Voermans J., Smits S.L., Osterhaus A.D., Al-Mawlawi N., Al-Romaihi H.E., AlHajri M.M., El-Sayed A.M., Mohran K.A., Ghobashy H., Alhajri F., Al-Thani M., Al-Marri S.A., El-Maghraby M.M., Koopmans M.P., Haagmans B.L. Isolation of MERS coronavirus from a dromedary camel, Qatar, 2014 // Emerg. Infect. Dis. 2014b. V. 20. № 8. P. 1339–1342. https://dx.doi.org/10.3201%2Feid2008.140663
Sabir J.S., Lam T.T., Ahmed M.M., Li L., Shen Y., Abo-Aba S.E., Qureshi M.I., Abu-Zeid M., Zhang Y., Khiyami M.A., Alharbi N.S., Hajrah N.H., Sabir M.J., Mutwakil M.H., Kabli S.A., Alsulaimany F.A., Obaid A.Y., Zhou B., Smith D.K., Holmes E.C., Zhu H., Guan Y. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia // Science. 2016. V. 351. № 6268. P. 81–84. https://doi.org/10.1126/science.aac8608
Samara E.M., Abdoun K.A. Concerns about misinterpretation of recent scientific data implicating dromedary camels in epidemiology of Middle East respiratory syndrome (MERS) // mBio. 2014. V. 5. № 4. P. e01430–01414. https://dx.doi.org/10.1128%2FmBio.01430-14
Sazmand A., Joachim A., Otranto D. Zoonotic parasites of dromedary camels: so important, so ignored // Parasit. Vectors. 2019. V. 12. № 1. P. 610. https://dx.doi.org/10.1186%2Fs13071-019-3863-3
Sekar A., Packyam M., Kim K. Growth enhancement of shrimp and reduction of shrimp infection by Vibrio parahaemolyticus and white spot syndrome virus with dietary administration of Bacillus sp. Mk22 // Microb. Biotechnol. Lett. 2016. V. 44. P. 261–267. https://doi.org/10.4014/mbl.1605.05001
Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., Zhao Y., Liu P., Liang L., Cui P., Wang J., Zhang X., Guan Y., Tan W., Wu G., Chen H., Bu Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2 // Science. 2020. P. eabb7015. https://dx.doi.org/10.1126%2Fscience.abb7015
Shi W., Li J., Zhou H., Gao G.F. Pathogen genomic surveillance elucidates the origins, transmission and evolution of emerging viral agents in China // Sci. China Life Sci. 2017. V. 60. № 12. P. 1317–1330. https://dx.doi.org/10.1007%2Fs11427-017-9211-0
So R.T.Y., Chu D.K.W., Miguel E., Perera R.A.P.M., Oladipo J.O., Fassi-Fihri O., Aylet G., Ko R.L.W., Zhou Z., Cheng M.S., Kuranga S.A., Roger F.L., Chevalier V., Webby R.J., Woo P.C.Y., Poon L.L.M., Peiris M. Diversity of Dromedary Camel Coronavirus HKU23 in African Camels Revealed Multiple Recombination Events among Closely Related Betacoronaviruses of the Subgenus Embecovirus // J. Virol. 2019. V. 93. № 23. P. 1–18. https://doi.org/10.1128/JVI.01236-19
Sun J., He W.T., Wang L., Lai A., Ji X., Zhai X., Li G., Suchard M.A., Tian J., Zhou J., Veit M., Su S. COVID-19: Epidemiology, evolution, and cross-disciplinary perspectives // Trends. Mol. Med. 2020. V. 26. № 5. P. 483–495. https://dx.doi.org/10.1016%2Fj.molmed.2020.02.008
Tang X., Wu C., Li X., Song Y., Yao X., Wu X., Duan Y., Zhang H., Wang Y., Qian Z., Cui J., Lu J. On the origin and continuing evolution of SARS-CoV-2 // Nat. Sci. Rev. 2020. nwaa036. https://doi.org/10.1093/nsr/nwaa036
Temmam S., Barbarino A., Maso D., Behillil S., Enouf V., Huon C., Jaraud A., Chevallier L., Backovic M., Pérot P., Verwaerde P., Tiret L., van der Werf S., Eloit M. Absence of SARS-CoV-2 infection in cats and dogs in close contact with a cluster of COVID-19 patients in a veterinary campus // bioRxiv. 2020. [Epub ahead of print]https://doi.org/10.1101/2020.04.07.029090
Tseng C.T., Sbrana E., Iwata-Yoshikawa N., Newman P.C., Garron T., Atmar R.L., Peters C.J., Couch R.B. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus // PLoS One. 2012. V. 7. № 4. P. e35421. https://doi.org/10.1371/journal.pone.0035421
Tu C., Crameri G., Kong X., Chen J., Sun Y., Yu M., Xiang H., Xia X., Liu S., Ren T., Yu Y., Eaton B.T., Xuan H., Wang L.F. Antibodies to SARS coronavirus in civets // Emerg. Infect. Dis. 2004. V. 10. № 12. P. 2244–2248. https://dx.doi.org/10.3201%2Feid1012.040520
van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D., Haagmans B.L., Gorbalenya A.E., Snijder E.J., Fouchier R.A. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans // mBio. 2012. V. 3. № 6. P. e00473-12. https://dx.doi.org/10.1128%2FmBio.00473-12
van den Brand J.M., Haagmans B.L., Leijten L., van Riel D., Martina B.E., Osterhaus A.D., Kuiken T. Pathology of experimental SARS coronavirus infection in cats and ferrets // Vet. Pathol. 2008. V. 45. № 4. P. 551–562. https://doi.org/10.1354/vp.45-4-551
Vijgen L., Keyaerts E., Moës E., Thoelen I., Wollants E., Lemey P., Vandamme A.M., Van Ranst M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event // J. Virol. 2005. V. 79. № 3. P. 1595–1604. https://dx.doi.org/10.1128%2FJVI.79.3.1595-1604.2005
Vijgen L., Keyaerts E., Lemey P., Maes P., Van Reeth K., Nauwynck H., Pensaert M., Van Ranst M. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43 // J. Virol. 2006. V. 80. № 14. P. 7270–7274. https://dx.doi.org/10.1128%2FJVI.02675-05
Wicker L.V., Canfield P.J., Higgins D.P. Potential Pathogens Reported in Species of the Family Viverridae and Their Implications for Human and Animal Health // Zoonoses Public Health. 2017. V. 64. № 2. P. 75–93. https://doi.org/10.1111/zph.12290
Widagdo W., Sooksawasdi Na Ayudhya S., Hundie G.B., Haagmans B.L. Host determinants of MERS-CoV transmission and pathogenesis // Viruses. 2019. V. 11. № 3. P. E280. https://doi.org/10.3390/v11030280
Wong A.C.P., Li X., Lau S.K.P., Woo P.C.Y. Global Epidemiology of bat coronaviruses // Viruses. 2019. V. 11. № 2. P. 174. https://dx.doi.org/10.3390%2Fv11020174
Woo P.C., Lau S.K., Fan R.Y., Lau C.C., Wong E.Y., Joseph S., Tsang A.K., Wernery R., Yip C.C., Tsang C.C., Wernery U., Yuen K.Y. Isolation and characterization of dromedary camel coronavirus UAE-HKU23 from dromedaries of the Middle East: minimal serological cross-reactivity between MERS coronavirus and dromedary camel coronavirus UAE-HKU23 // Int. J. Mol. Sci. 2016. V. 17. № 5. P. 691. https://dx.doi.org/10.3390%2Fijms17050691
Woo P.C., Lau S.K., Wernery U., Wong E.Y., Tsang A.K., Johnson B., Yip C.C., Lau C.C., Sivakumar S., Cai J.P., Fan R.Y., Chan K.H., Mareena R., Yuen K.Y. Novel betacoronavirus in dromedaries of the Middle East, 2013 // Emerg. Infect. Dis. 2014. V. 20. № 4. P. 560–572. https://dx.doi.org/10.3201%2Feid2004.131769
Wu D., Tu C., Xin C., Xuan H., Meng Q., Liu Y., Yu Y., Guan Y., Jiang Y., Yin X., Crameri G., Wang M., Li C., Liu S., Liao M., Feng L., Xiang H., Sun J., Chen J., Sun Y., Gu S., Liu N., Fu D., Eaton B.T., Wang L.F., Kong X. Civets are equally susceptible to experimental infection by two different severe acute respiratory syndrome coronavirus isolates // J. Virol. 2005. V. 79. № 4. P. 2620–2625. https://dx.doi.org/10.1128%2FJVI.79.4.2620-2625.2005
Xia X. Extreme genomic CpG deficiency in SARS-CoV-2 and evasion of host antiviral defense // Mol. Biol. Evol. msaa094. 2020. [Epub ahead of print]https://doi.org/10.1093/molbev/msaa094
Xiao Y., Meng Q., Yin X., Guan Y., Liu Y., Li C., Wang M., Liu G., Tong T., Wang L.F., Kong X., Wu D. Pathological changes in masked palm civets experimentally infected by severe acute respiratory syndrome (SARS) coronavirus // J. Comp. Pathol. 2008. V. 138. № 4. P. 171–179. https://dx.doi.org/10.1016%2Fj.jcpa.2007.12.005
Yang Y., Peng F., Wang R., Guan K., Jiang T., Xu G., Sun J., Chang C. The deadly coronaviruses: The 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China // J. Autoimmun. 2020. V. 109. P. 102434. https://dx.doi.org/10.1016%2Fj.jaut.2020.102434
Ye Z.W., Yuan S., Yuen K.S., Fung S.Y., Chan C.P., Jin D.Y. Zoonotic origins of human coronaviruses // Int. J. Biol. Sci. 2020. V. 16. № 10. P. 1686–1697. https://dx.doi.org/10.7150%2Fijbs.45472
Zhang G., Cowled C., Shi Z., Huang Z., Bishop-Lilly K.A., Fang X., Wynne J.W., Xiong Z., Baker M.L., Zhao W., Tachedjian M., Zhu Y., Zhou P., Jiang X., Ng J., Yang L., Wu L., Xiao J., Feng Y., Chen Y., Sun X., Zhang Y., Marsh G.A., Crameri G., Broder C.C., Frey K.G., Wang L.F., Wang J. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity // Science. 2013. V. 339. № 6118. P. 456–460. https://doi.org/10.1126/science.1230835
Zhang Q., Zhang H., Huang K., Yang Y., Hui X., Gao J., He X., Li C., Gong W., Zhang Y., Peng C., Gao X., Chen H., Zou Z., Shi Z., Jin M. SARS-CoV-2 neutralizing serum antibodies in cats: a serological investigation // bioRxiv. 2020. [Epub ahead of print]https://doi.org/10.1101/2020.04.01.021196
Zhao S., Li W., Schuurman N., van Kuppeveld F., Bosch B.J., Egberink H. Serological screening for coronavirus infections in cats // Viruses. 2019. V. 11. № 8. P. 743. https://dx.doi.org/10.3390%2Fv11080743
Zhu S., Zimmerman D., Deem S.L. A review of zoonotic pathogens of dromedary camels // Ecohealth. 2019. V. 16. № 2. P. 356–377. https://doi.org/10.1007/s10393-019-01413-7
Дополнительные материалы отсутствуют.
Инструменты
Известия РАН. Серия биологическая


