Молекулярная биология, 2022, T. 56, № 2, стр. 227-243
Регулом микроРНК при различных фенотипах атеросклероза
М. С. Назаренко a, b, *, Ю. А. Королёва a, А. А. Зарубин a, А. А. Слепцов a
a Научно-исследовательский институт медицинской генетики, Томский национальный исследовательский медицинский центр Российской академии наук
634050 Томск, Россия
b Сибирский государственный медицинский университет
634050 Томск, Россия
* E-mail: maria.nazarenko@medgenetics.ru
Поступила в редакцию 29.07.2021
После доработки 25.08.2021
Принята к публикации 26.08.2021
- EDN: ZUEBCV
- DOI: 10.31857/S0026898422020136
Аннотация
Нарушение регуляции экспрессии микроРНК связано с предрасположенностью ко многим заболеваниям, в том числе к атеросклеротическому поражению коронарных и сонных артерий и развитию таких осложнений, как ишемическая болезнь сердца, инфаркт миокарда, хроническая ишемия головного мозга, ишемический инсульт. В последнее время появляется все больше работ, в которых анализируется регулом микроРНК, включающий сеть регуляторных элементов экспрессии собственно микроРНК и мишеней, находящихся под их контролем. В обзоре рассмотрена экспрессия микроРНК и изменения метилирования ДНК в области генов микроРНК в артериях человека при их атеросклеротическом поражении, а также проанализирована связь однонуклеотидных полиморфизмов и вариаций числа копий участков ДНК в области генов микроРНК с клиническими осложнениями атеросклероза.
ВВЕДЕНИЕ
Атеросклероз – комплексное, хроническое воспалительное заболевание с прогредиентным течением, характеризующееся ремоделированием артерий, появлением в них атеросклеротических бляшек со стенозом просвета и развитием ишемии, тромбоза/эмболии органов-мишеней, в частности сердца, головного мозга и др. Патологические изменения в артериях включают дисфункцию эндотелиальных клеток, отложение окисленных липопротеинов и инфильтрацию стенки артерии моноцитами/макрофагами с их превращением в пенистые клетки, утолщение интимы, миграцию и пролиферацию гладкомышечных клеток (ГМК), трансдифференцировку фенотипа клеток, кальцификацию, ангиогенез, образование фиброзной бляшки с липидно-некротическим ядром и последующей ее дестабилизацией [1–3].
Практически во всех стадиях развития атеросклероза (от ранней дисфункции эндотелия до эрозии или разрыва нестабильной атеросклеротической бляшки) участвуют микроРНК – малые регуляторные эволюционно консервативные некодирующие РНК (~16–27 н.) [4–8]. Экспрессия микроРНК в клетках крови и артерий на каждой стадии развития атеросклероза контролируется различными стимулами, а сами микроРНК регулируют множество сигнальных путей, вовлеченных в формирование и дестабилизацию атеросклеротической бляшки [5, 6].
Механизм действия микроРНК заключается преимущественно в негативной регуляции экспрессии генов через деградацию мРНК или репрессию трансляции мРНК-мишеней в цитоплазме клеток [9–11]. Однако в клеточном ядре отдельные микроРНК могут активировать транскрипцию генов, действуя как триггеры энхансеров [11].
Согласно онлайн-базе данных miRBase v. 22.1, в тканях человека обнаружено более 2600 микроРНК [12], которые могут регулировать экспрессию более 60% всех белоккодирующих генов, участвуя тем самым во всех основных биологических процессах [9, 12, 13]. Транскрипция генов микроРНК, локализованных в межгенном пространстве, контролируется их собственными регуляторными элементами, в то время как микроРНК, расположенные в области белоккодирующих генов, часто обладают общим промотором с геном-хозяином [14, 15]. Тем не менее, примерно 50% внутригенных микроРНК имеют собственный промотор [16].
Считается, что гены микроРНК составляют около 1–5% нуклеотидной последовательности всех генов человека [11, 17]. Гены микроРНК могут располагаться либо в области белоккодирующих генов (экзонов/интронов), либо в межгенных областях [14, 18]. Согласно базе данных miRIAD [19], среди 1881 микроРНК число экзонных микроРНК составляет 169 (9%), интронных – 988 (52.5%), а межгенных – 724 (38.5%).
В базе данных HMDD v. 3.0: the Human microRNA Disease Database (обновление от 27 марта 2019) [20] представлена информация о 116 экспериментально подтвержденных микроРНК, связанных с атеросклерозом, 26 микроРНК – с атеросклерозом сонных артерий и 67 – с ишемическим инсультом, 85 – с ишемической болезнью сердца (ИБС) и 60 – с острым коронарным синдромом и инфарктом миокарда.
Цель настоящего обзора заключалась в обобщении результатов изучения регулома микроРНК, включающего сеть регуляторных элементов экспрессии собственно микроРНК и мишеней, находящихся под их контролем при атеросклерозе сонных и коронарных артерий.
Представленный обзор сфокусирован на исследовании молекулярно-генетических механизмов атеросклероза через дисрегуляцию микроРНК в клетках и органах-мишенях заболевания (атеросклеротические бляшки), что отличает его от обзоров, в которых охарактеризованы пациенты с острыми сосудистыми событиями и проведен поиск биомаркеров данных патологических состояний через определение экспрессии микроРНК в плазме/сыворотке [21].
Связь микроРНК с атеросклерозом рассмотрена на уровнях экспрессии микроРНК, изменения метилирования ДНК в области генов микроРНК и ДНК-полиморфизма в области генов микроРНК. Сопоставление полученных при этом данных позволит охарактеризовать регулом микроРНК при различных фенотипах атеросклероза у человека (ИБС, инфаркт миокарда, хроническая ишемия головного мозга, ишемический инсульт), что поможет более точно планировать проведение экспериментальных работ, направленных на понимание функций микроРНК и молекулярных механизмов регуляции их функциональной активности, с использованием культур клеток и модельных животных.
ЭКСПРЕССИЯ микроРНК В КОРОНАРНЫХ И СОННЫХ АРТЕРИЯХ ПРИ ИХ АТЕРОСКЛЕРОТИЧЕСКОМ ПОРАЖЕНИИ
К настоящему времени опубликовано несколько десятков работ, в которых с использованием различных групп сравнения и методов проанализировано изменение экспрессии микроРНК в цельных сосудах человека при их поражении атеросклерозом. В большинстве исследований анализировали экспрессию отдельных микроРНК в тканях артерий с помощью ПЦР в режиме реального времени [22–33]. Кроме того, сравнивали экспрессию микроРНК в атеросклеротических бляшках коронарных или сонных артерий и в интактных сосудах, используя для этого микрочипы [34–40].
В частности, в пионерском исследовании Raitoharju E. и соавт. (2011) с использованием микрочипов выявлено 58 микроРНК, которые дифференциально экспрессировались в атеросклеротических бляшках различной локализации по сравнению с интактными внутренними грудными артериями [34]. Увеличение экспрессии пяти микроРНК (miR-21, -34a, -146a, -146b-5p и -210) в атеросклеротических бляшках подтверждено с помощью ПЦР в режиме реального времени на большей по размеру выборке тканей артерий. Экспрессия 187 мРНК генов-мишеней данных микроРНК была снижена в атеросклеротических бляшках. Белковые продукты этих генов участвуют в передаче сигнала, регуляции транскрипции и транспорте везикул.
В табл. 1 представлена 31 микроРНК, экспрессия которых в атеросклеротических бляшках коронарных и сонных артерий статистически значимо отличается от экспрессии в интактных тканях сосудов, либо между нестабильными (симптоматическими) и стабильными (асимптоматическими) атеросклеротическими бляшками в двух или более исследованиях [22–24, 26–41].
Таблица 1.
Экспрессия микроРНК в атеросклеротических бляшках коронарных и сонных артерий
| miR | Сонные артерии | Коронарные артерии | Ссылка |
|---|---|---|---|
| Атеросклеротическая бляшка vs интактные ткани/нестабильная vs стабильная атеросклеротическая бляшка | Атеросклеротическая бляшка vs интактные ткани/нестабильная vs стабильная атеросклеротическая бляшка | ||
| -1† | ↓ | ↓ | [37–39] |
| let-7f† | ↓ | ↓ | [38, 39] |
| -9 | НД | ↑ | [37] |
| НД | ↓ | [39] | |
| -10a | НД | ↑ | [37] |
| НД | ↓ | [40] | |
| -10b | НД | ↑ | [37] |
| НД | ↓ | [40] | |
| -16† | НД | ↑ | [37] |
| НД | ↓ | [40] | |
| -19a | НД | ↓ | [37, 40] |
| -19b | ↑ | ↑ | [37–39] |
| НД | ↓ | [40] | |
| -21† | ↑ | ↑ | [28, 34, 38, 39] |
| ↓ К (≤6 недель) | ↓ | [37, 38] | |
| ↓ К (>14 дней), Г (тонкая покрышка, разрывы) | НД | [33] | |
| -22 | ↑ | ↓ | [37–39] |
| -24† | НД | ↓ Г | [26] |
| НД | ↓ | [37] | |
| -25 | НД | ↑ | [37] |
| НД | ↓ | [40] | |
| -29b† | ↑ | ↑ | [34, 39] |
| ↓ | ↓ | [37, 38] | |
| -29c | НД | ↓ | [37] |
| НД | ↑ | [36] | |
| -34a | ↑ | ↓ | [34, 37] |
| -92a | ↓ | ↓ | [34, 37] |
| НД | ↑ | [39] | |
| -100 | ↑ К | ↑ | [22, 29, 36] |
| НД | ↓ | [37] | |
| -106b† | НД | ↓ | [37, 40] |
| -125a | ↑ К | ↓ | [29, 41] |
| -127 | ↑ К | НД | [22, 29] |
| -133a† | ↑ К | НД | [22, 29] |
| -133b | ↑ К | ↓ | [22, 37] |
| ↑ Г (низкая степень кальцификации) | НД | [42] | |
| -143 | ↓ | ↓ | [34, 37, 39] |
| ↑ | НД | [38] | |
| ↓ К (≤6 недель) | НД | [38] | |
| -145† | ↓ | ↓ | [23, 37] |
| ↑ К | НД | [22, 29] | |
| -146a† | ↑ | ↑ | [28, 34, 37] |
| -150 | НД | ↑ | [32, 37] |
| -155† | ↑ | ↓ | [24, 34, 36] |
| НД | ↑ | [41] | |
| -221 | ↑ К | ↑ | [24, 29, 31, 36, 37] |
| ↓ К (<5 дней) | НД | [24] | |
| -223† | НД | ↑ | [37, 39] |
| -486† | НД | ↑ | [37] |
| НД | ↓ | [40] | |
| -497 | НД | ↑ | [36] |
| НД | ↓ | [37] |
Примечание. Атеросклеротическая бляшка, нестабильная клинически (К) и гистологически (Г). НД – нет данных. † – микроРНК, которые определяются в сыворотке/плазме крови индивидов при ИБС, остром коронарном синдроме, инфаркте миокарда, ишемическом и геморрагическом инсультах (согласно [21]).
Всего в коронарных и сонных артериях при их атеросклеротическом поражении зарегистрировано изменение экспрессии 31 микроРНК (табл. 1). В сонных артериях преобладали микроРНК, экспрессия которых в атеросклеротических бляшках увеличена по сравнению с контролем (miR-19b, -22, -34a, -100, -125a, -127, -133a, -133b, -146a, -155). Экспрессия let-7f, miR-1 и miR-92a снижена в атеросклеротически измененных сонных артериях, а для miR-21, -29b, -143, -145, -221 получены противоречивые результаты в разных исследованиях.
Коронарные артерии, наоборот, отличались большим количеством микроРНК со снижением экспрессии (let-7f, miR-1, -19a, -22, -24, -34a, -106b, -125a, -133b, -143, -145) и разнонаправленным ее изменением в разных исследованиях (miR-9, -10a, -10b, -16, -19b, -21, -25, -29b, -29c, -92a, -100, -155, -486, -497). Увеличение экспрессии miR-146a, -150, -221, -223 зарегистрировано в атеросклеротических бляшках коронарных артерий.
В коронарных и сонных артериях выявлены однонаправленные изменения экспрессии miR-146a (увеличение), miR-1 и let-7f (уменьшение), тогда как экспрессия miR-21 и miR-29b в разных исследованиях или увеличивалась, или уменьшалась.
В сонных артериях экспрессия miR-21, -100, ‑125a, -127, -133a/b, -143/-145, -221 связана с нестабильностью атеросклеротических бляшек [22, 27, 29, 33, 38, 42]. Нестабильные бляшки характеризуются увеличением экспрессии большинства микроРНК (miR-100, -127, -133a/b, -145) и снижением экспрессии miR-21 и miR-143. Противоречивые результаты получены для miR-221. В коронарных артериях снижение экспрессии miR-24 связано с нестабильностью атеросклеротической бляшки [26].
Экспрессию микроРНК оценивали в биоптатах атеросклеротических бляшек сонных артерий, полученных при эндартерэктомии. В большинстве исследований использовали клинический критерий нестабильности атеросклеротических бляшек – пациенты с симптоматическим стенозом сонных артерий, которые имели острое нарушение мозгового кровообращения (транзиторные ишемические атаки/ипсилатеральный инсульт) менее чем за 6 мес. до операции. Однако в отдельных исследованиях сроки от острого нарушения мозгового кровообращения до операции варьировали от менее 5 дней и до 3 мес. [27, 33, 38]. Лишь в единичных работах экспрессию микроРНК определяли в макрофагах, взятых из областей гистологически нестабильной атеросклеротической бляшки (с низкой степенью кальцификации [42]), в фиброзной покрышке атеросклеротических бляшек с истончением (<200 мкм) и разрывами [33].
Показана связь экспрессии микроРНК miR-145 и семейства let-7 в атеросклеротических бляшках сонных артерий с факторами риска атеросклероза (артериальная гипертензия и сахарный диабет типа 2) [25, 43].
В систематическом обзоре Sharma и соавт. отмечено, что 155 микроРНК, определяемых в сыворотке/плазме крови индивидов, ассоциированы с ИБС, острым коронарным синдромом, инфарктом миокарда, ишемическим и геморрагическим инсультами [21]. Из них 33 микроРНК обнаружены более чем в одном исследовании. Сопоставление этих микроРНК в сыворотке/плазме и в тканях коронарных и сонных артерий при их атеросклеротическом поражении выявило среди них 13 общих (let-7f, miR-1, -16, -21, -24, -29b, -106b, -133a, -145, -146a, -155, -223, -486; табл. 1). Эти микроРНК, по-видимому, представляют особый интерес не только в качестве биомаркеров осложненного течения атеросклероза, но и как терапевтические мишени при данной патологии.
Как правило, в атеросклеротических бляшках коронарных и сонных артерий анализируют экспрессию отдельных микроРНК. Принимая во внимание сложный и длительный процесс формирования атеросклеротических бляшек, характеризующихся высокой клеточной гетерогенностью, можно предположить возможность совместного изменения экспрессии различных микроРНК при развитии атеросклероза или изменения клеточного состава артерий с формированием модулей коэкспрессии микроРНК. Однако многие аспекты взаимосвязей данных молекул между собой и регуляции мРНК их генов-мишеней остаются малоизученными. Существенную помощь в решении данной проблемы оказывают биоинформатические подходы, инструменты и базы данных, которые позволяют функционально аннотировать микроРНК и их гены-мишени [44, 45].
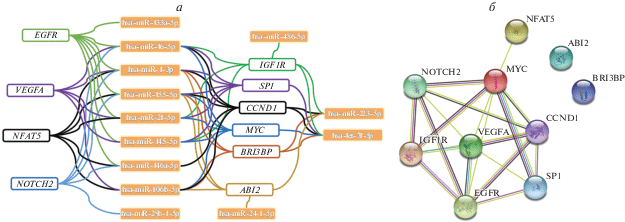
Например, характеристика 13 упомянутых микроРНК (let-7f, miR-1, -16, -21, -24, -29b, -106b, -133a, -145, -146a, -155, -223, -486), дифференциально экспрессирующихся в коронарных/сонных артериях, и определяемых в сыворотке/плазме крови индивидов с клиническими осложнениями атеросклероза с помощью инструментов miRTargetLink 2.0 [44], miEAA 2.0 [46] и базы miRTarBase [47], позволяет выделить 10 генов-мишеней (EGFR, ABI2, IGF1R, NFAT5, BRI3BP, VEGFA, CCND1, SP1, MYC, NOTCH2), которые регулируются пятью и более микроРНК (рис. 1а). Гены EGFR и CCND1 служат мишенями семи микроРНК, а их белковые продукты взаимодействуют с IGF1R, VEGFA, SP1, MYC, NOTCH2 и контролируют ангиогенез (рис. 1б). Активация иммунного ответа связана с белковым продуктом гена NFAT5, миграция и адгезия клеток – ABI2, апоптоз – BRI3BP.
Рис. 1.
Взаимодействие микроРНК и их генов мишеней при атеросклерозе коронарных и сонных артерий. а – МикроРНК и их гены-мишени, которые регулируются пятью и более микроРНК; б – взаимодействие между собой белковых продуктов генов-мишеней микроРНК. Для визуализации использован инструмент STRING v. 11.0 [48].
Следует отметить, что методические подходы к изучению экспрессии микроРНК в тканях артерий при их поражении атеросклерозом с помощью микрочипов и ПЦР в режиме реального времени имеют определенные недостатки, так как анализируют только уже известные микроРНК. Кроме того, определенную сложность представляет сравнение результатов, полученных в разных работах, что связано с несопоставимостью разных платформ микрочипов или групп сравнения. Массовое параллельное секвенирование микроРНК (microRNA-seq) свободно от таких недостатков, но на данный момент этот подход не применяли для исследования клеток атеросклеротической бляшки, его использовали только для анализа экспрессии микроРНК в плазме или в цельной крови пациентов с ИБС [49].
МЕТИЛИРОВАНИЕ ДНК В ОБЛАСТИ ГЕНОВ микроРНК В КОРОНАРНЫХ И СОННЫХ АРТЕРИЯХ ПРИ ИХ АТЕРОСКЛЕРОТИЧЕСКОМ ПОРАЖЕНИИ
Несмотря на то что микроРНК участвуют в эпигенетической регуляции функциональной активности многих генов, сами эти молекулы могут быть мишенями для эпигенетических модификаций. Экспрессия микроРНК контролируется, в том числе, посредством эпигенетических механизмов, один из которых – метилирование CpG-островков (скоплений CpG-динуклеотидов), перекрывающих промоторные участки генов или локализованных рядом с сайтом начала транскрипции микроРНК [15, 50]. Обычно CpG-островки гипометилированы, а их избыточное гиперметилирование подавляет транскрипцию и, следовательно, ингибирует экспрессию соответствующих генов [51–53].
Метилирование более характерно для подавления экспрессии генов микроРНК, чем белоккодирующих генов. По данным разных авторов метилированию подвержено 11–30% генов микроРНК и только 1–2% белоккодирующих генов [15, 50, 54]. Кроме того, примерно 50% генов микроРНК содержат CpG-островки и, следовательно, могут регулироваться посредством метилирования ДНК [55, 56].
Однако регуляция экспрессии микроРНК путем метилирования ДНК происходит и вне CpG-островков [55]. В частности, контроль экспрессии интронных микроРНК может осуществляться в области их собственных промоторов и обладать тканевой и клеточной специфичностью. Собственные промоторы интронных микроРНК представляют собой участки генома с низким содержанием CpG-динуклеотидов, насыщенные такими элементами, как TATA-боксы и сайты связывания различных факторов транскрипции [16]. Кроме того, изменение метилирования ДНК в области энхансеров также может модулировать экспрессию микроРНК [57].
При исследовании атеросклеротических поражений артерий различной локализации внимание уделяется, главным образом, анализу метилирования ДНК в области промоторов белоккодирующих генов. Лишь в нескольких работах изменение уровня метилирования ДНК выявлено в области генов микроРНК в атеросклеротических бляшках [58–63].
В частности, с использованием микрочипа Illumina Infinium Human Methylation27 BeadChip выявлено снижение уровня метилирования в области CpG-островка (расположенного в области промотора гена HOXD4), на который приходится кодирующий регион гена MIR10B, в тканях атеросклеротических бляшек коронарных и сонных артерий, а также в лейкоцитах периферической крови больных атеросклерозом по сравнению с непораженными внутренними грудными артериями и большими подкожными венами нижних конечностей [58, 62].
Следует отметить, что экспрессия miR-10b изменяется в цельных артериях человека при их поражении атеросклерозом [37, 40]. У мышей с нокаутом гена Mir10b, получавших богатую холестерином диету, и с частичным лигированием сонных артерий, атеросклероз не развивался [64]. Как показано на клеточных культурах, увеличение экспрессии miR-10b в эндотелиальных клетках (ЭК), действующей через гены-мишени LTBP1 и HOXD10, способствует их миграции и ангиогенезу, а также снижению способности ЭК типа II подавлять пролиферацию ГМК. В макрофагах увеличение экспрессии miR-10b через гены-мишени ABCA1 и ABCG1 приводит к уменьшению эффлюкса и обратного транспорта холестерина, а при воздействии данной микроРНК на ген-мишень TIP30 усиливается пролиферация ГМК [64–67].
Китайские исследователи оценивали уровень метилирования ДНК, экспрессию мРНК и микроРНК в пуллированных post mortem образцах коронарных артерий из области выраженных атеросклеротических бляшек и интактных регионов, используя микрочип GoldenGate Methylation Cancer Panel I (“Illumina”, США), 22K human genome array chip (“CapitalBio Corp”, США) и miRCURY LNA Array (“Exiqon”, США). Выявлены не только гены, клеточные процессы и сигнальные пути, участие которых в атеросклерозе рассматривали ранее, но и miR-519d [59].
В другой работе с использованием микрочипа Illumina HumanMethylation450 BeadChip сравнили уровень метилирования отдельных CpG-сайтов в атеросклеротических бляшках аорты и сонных артерий и в непораженных тканях аорты. Причем образцы аорты взяты от одних и тех же индивидов post mortem, а образцы сонных артерий, полученные в результате каротидной эндатерэктомии, представляли собой стабильные бляшки морфогистологических типов III–VI. Среди генов, дифференциально метилированных в атеросклеротических бляшках и в непораженной части аорты, выявлен ген MIR23B. Изменение уровня метилирования ДНК в области этого гена и его экспрессии в артериях подтверждено с помощью альтернативных методов [60].
Финские исследователи с помощью технологии полногеномного бисульфитного секвенирования оценили уровень и рисунок метилирования ДНК в атеросклеротических бляшках бедренных артерий и интактных образцах внутренних грудных артерий, а также сопоставили их с данными экспрессионных микрочипов [61]. Более 140 областей генов микроРНК были гипометилированы в атеросклеротических бляшках, включая связанные с атеросклерозом микроРНК miR-10b, -27b и -758. Существенное гипометилирование выявлено в локусе 14q32, содержащем крупный кластер генов микроРНК. Повышение экспрессии генов этого кластера (miR-127, -136, -410, -431, -432 и -433) показано также в атеросклеротических бляшках. Таким образом, это одно из первых исследований, в котором установлена связь между метилированием ДНК и экспрессией микроРНК.
Однако в одной из недавних работ обнаружены значительные различия в экспрессии кластера микроРНК, расположенных в локусе 14q32, в здоровых тканях сосудов различной локализации (n = 109). Уровень метилирования ДНК в трех регионах CpG-островков (дифференциально метилированные регионы – DMR) в данном локусе также сильно варьирует между разными сосудами и ассоциирован с их патологическими изменениями, но не связан с экспрессией при-микроРНК и зрелых микроРНК [68].
В работе Ehrlich и соавт. с помощью полногеномного бисульфитного секвенирования трех образцов аорты выявлено гиперметилирование энхансерной области гена CARMN (MIR143HG) протяженностью 26 т.п.н. в пораженной атеросклерозом аорте по сравнению с непораженной аортой и другими тканями, а также потенциальная связь изменения метилирования ДНК анализируемой области с экспрессией miR-143 и miR-145 [63].
В то же время, при анализе ассоциации экспрессии 283 микроРНК с метилированием более чем 400000 CpG-сайтов в клетках цельной крови, полученных от 3565 пациентов с ИБС в исследовании Framingham Heart Study, обнаружена связь дифференциального метилирования 227 CpG-сайтов (cis-eQTM) с экспрессией 40 близлежащих микроРНК на уровне значимости p < 0.01 [69]. Чаще всего cis-eQTM располагались в промоторных регионах и областях с Polycomb-репрессированным состоянием; для 60% из них показана обратная ассоциация метилирования с экспрессией близлежащей микроРНК. При проведении Менделевской рандомизации дополнительно выявили 58 пар cis-eQTM CpG-сайтов и микроРНК, изменения метилирования ДНК в которых, по-видимому, влияют на экспрессию микроРНК. В число этих cis-eQTM входит cg18089426, влияющий на экспрессию miR-127. Однако повышение уровня метилирования cis-eQTM в областях “тела” генов микроРНК, напротив, приводило к усилению транскрипции. В частности, обнаружена положительная связь метилирования ДНК и экспрессии cg06000878 и miR-100, а также cg11682508 и miR-133a [69].
Таким образом, изменения уровня метилирования ДНК в области генов микроРНК (miR-10b, miR-23b, локус 14q32 (miR-127, -136, -410, -431, -432 и -433), miR-143 и miR-145) в клетках атеросклеротических бляшек анализировали в немногочисленных работах, выполненных на цельных и разных по своей локализации сосудах. Остается неясным, специфичен ли профиль метилирования ДНК, в том числе в области генов микроРНК, в артериях различной локализации, а также связан ли он с клеточным составом и клиническими признаками атеросклероза. Кроме того, оценку роли изменений метилирования ДНК в области генов микроРНК в пораженных атеросклерозом артериях у человека затрудняет динамический характер заболевания, а также высокая клеточная гетерогенность тканей сосудов. В связи с этим проведение таких исследований необходимо дополнять использованием модельных животных и клеточных культур.
ПОЛИМОРФИЗМ ДНК В ОБЛАСТИ ГЕНОВ микроРНК ПРИ ЗАБОЛЕВАНИЯХ, АССОЦИИРОВАННЫХ С АТЕРОСКЛЕРОЗОМ
Широкогеномные исследования ассоциаций показали, что большая часть однонуклеотидных полиморфизмов (SNP), ассоциированных с атеросклерозом артерий, локализована в некодирующих регионах генома и/или в регуляторных элементах [70]. Учитывая, что генетическая предрасположенность играет важную роль в риске развития данных патологий, полиморфизм в генах микроРНК (в регуляторных регионах, в при- и пре-микроРНК, в зрелой микроРНК), генах их биогенеза и генах-мишенях может иметь функциональную значимость для формирования патологии [71].
Предполагается, что количество полиморфных вариантов в генах микроРНК, особенно в генах их биогенеза, меньше, чем в генах-мишенях, поскольку микроРНК консервативны и имеют небольшой размер. В нуклеотидной последовательности 1075 пре-микроРНК выявлено 2257 SNP, в области узнавания мРНК-мишени (seed-region) 207 микроРНК – 246 SNP [72]. В то же время, около 180 000 SNP расположены в 3′-нетранслируемой области (UTR) белоккодирующих генов, т.е. теоретически большое количество данных генетических вариантов может быть связано с микроРНК через мРНК [73].
К настоящему времени изучены ассоциации полиморфизма ДНК в области генов микроРНК с клиническими осложнениями атеросклероза коронарных и сонных артерий – ИБС и инфарктом миокарда, ишемическим инсультом, а также с рестенозом коронарных артерий (табл. 2). Наибольшее количество работ посвящено rs2910164:G > C (MIR146A) и rs3746444:A > G (MIR499A; MIR499B).
Таблица 2.
Ассоциация полиморфизма ДНК в области генов микроРНК с клиническими осложнениями атеросклероза коронарных и сонных артерий
| miR | SNP | Локализация SNP* | Аллель/ генотип риска |
Фенотип атеросклероза |
Популяция | Размер выборки (случай/контроль) | Ссылка |
|---|---|---|---|---|---|---|---|
| let-7 | rs13293512 | 8.5 т.п.н. выше гена MIRLET7A1 | TT | ИИ | Китай | 329/357 | [74] |
| -27a | rs895819 | В теле гена MIR27A | GG | ИМ | Китай | 287/646 | [75] |
| -146a | rs2910164 | В теле гена MIR146A | G/GG | Атеросклероз коронарных, сонных и периферических артерий | Турция | 150/145 | [76] |
| CC | Риск ССЗ – в Южной Корее и Индии, но защитный эффект в популяции Китая | Мета-анализ (Азия) | 5433/6278 (12 исследований) | [77] | |||
| C/ GC + CC | ИБС | Китай | 295/283 | [78] | |||
| CC | ИБС | Мета-анализ (разные страны) | 13186/14497 (30 исследований) | [79] | |||
| CC | ИБС | Мета-анализ (разные страны) | 3138/3097 (8 исследований) | [80] | |||
| CC | Рестеноз коронарных артерий | Германия | 101/105 | [81] | |||
| СС | Атеросклероз сонных артерий | Китай | 596/379 | [82] | |||
| C/CC | ИИ при атеросклерозе крупных артерий | Китай | 368/381 | [83] | |||
| C | ИИ | Китай | 297/300 | [84] | |||
| G | ИИ | Южная Корея | 678/553 | [85] | |||
| G/GG | Риск ИИ только для Южной Кореи | Мета-анализ (разные страны) | 3138/3097 (8 исследований) | [80] | |||
| G/GG | Риск ИИ только для Южной Кореи | Мета-анализ (Азия) | 2254/2506 (5 исследований) | [86] | |||
| GG | Повторный ИИ | Китай | 1139/1585 | [87] | |||
| -149 | rs2292832 | В теле гена MIR149 | T/TC + CC | ИБС | Южная Корея | 522/535 | [88] |
| -196a2 | rs11614913 | В теле гена MIR196A2 | CC+CT | Осложнения ИБС | Китай | 956/620 | [89] |
| C | ИМ | Россия | 325/185 (исходная выборка), 203/280 (валидационная выборка) | [90] | |||
| TT | Риск ИИ только в Китае | Мета-анализ (Азия) | 1873/1856 (4 исследования) | [86] | |||
| -200b | rs7549819 | 1.5 т.п.н. выше гена MIR200B | TT+TC | ИИ | Южная Корея | 523/400 | [91] |
| -423 | rs6505162 | В теле гена MIR423 | A/CA | ИБС | Индия | 100/117 | [92] |
| -499 | rs3746444 | В теле гена MIR499A и MIR499B | A | Атеросклероз коронарных, сонных и периферических артерий | Турция | 150/145 | [76] |
| GG | ИМ | Китай | 919/889 | [93] | |||
| G | ИИ | Иран | 470/489 | [94] | |||
| G | ИИ | Китай | 296/391 | [95] | |||
| GG | Риск ИИ только в китайской популяции | мета-анализ (Азия) | 1505/1475 (3 исследования) | [86] | |||
| -618 | rs2682818 | В теле гена MIR618 | GT+TT | Повторный ИИ | Китай | 74/779 | [96] |
| -4513 | rs2168518 | В теле гена MIR4513 | A | ИБС | Мета-анализ (CARDIoGRAMplusC4D, GWAS) | 63746/130681 | [97] |
Ассоциацию полиморфного варианта rs2910164:G > C (MIR146A) с атеросклерозом коронарных артерий изучали, главным образом, на китайских популяциях (табл. 2). В отдельных исследованиях установлено повышение риска ИБС и ишемического инсульта у носителей генотипа СС и аллеля С этого полиморфизма. Однако мета-анализ результатов, полученных на выборках жителей Азии, выявил межпопуляционные различия в ассоциации данного полиморфизма с разными патологическими фенотипами атеросклероза коронарных и сонных артерий [77, 79, 80, 86]. Не исключено также существование различий в молекулярно-генетических механизмах формирования атеросклеротического поражения коронарных и сонных артерий.
В единственной работе, проведенной на европейской выборке (жители Германии), показана связь генотипа СС rs2910164 c риском развития рестеноза коронарных артерий, а не собственно их атеросклеротического поражения [81]. Поскольку рестеноз представляет собой четко очерченный фенотип, который отличается от атеросклероза быстрым темпом развития, предполагается, что эпигенетические механизмы могут более тесно участвовать в патогенезе рестеноза по сравнению с атеросклерозом. С другой стороны, учитывая, что частота аллеля C rs2910164 в европейских популяциях ниже, чем у жителей Азии (ExAC 0.22 против 0.39 [98]), полученный результат может быть связан с недостаточной статистической мощностью исследования.
Аллель G и генотип GG rs3746444:A>G (MIR499A; MIR499B) оказались ассоцированными с развитием инфаркта миокарда и ИИ в популяциях Китая, Южной Кореи и Ирана (табл. 2). Однако в турецкой популяции риск развития атеросклероза коронарных, сонных и периферических артерий был выше у носителей аллеля A rs3746444. Противоречия в результатах исследований могут быть связаны со многими факторами, включая, прежде всего, небольшой размер выборки и недостаточную статистическую мощность (в последней работе), а также различия в этническом происхождении индивидов и их отягощенности коморбидными заболеваниями.
Функциональные последствия носительства полиморфного варианта будут различаться в зависимости от его расположения относительно генов микроРНК. Равномерное изменение общего количества всех микроРНК и, соответственно, экспрессии их генов-мишеней происходит при локализации SNP в генах биогенеза микроРНК. Однако к настоящему времени нет крупномасштабных работ, которые анализируют ассоциации полиморфизма ДНК в генах биогенеза микроРНК с сердечно-сосудистыми заболеваниями, обусловленными атеросклерозом.
Наиболее сильно на связывание микроРНК со своими мРНК-мишенями влияют SNP, локализующиеся в “seed-region” зрелой микроРНК. Эти варианты определяют усиление или ослабление связи микроРНК с мРНК, а также возможность потери связи микроРНК со своими изначальными мишенями и/или взаимодействия с новыми мишенями. Например, rs2168518:G > A, который располагается в “seed-region” miR-4513, ассоциирован с разными кардиометаболическими фенотипами (уровни глюкозы и липидов в сыворотке крови, артериальное давление), а также с ИБС [97, 99]. Показано, что аллель A rs2168518 снижает уровень и активность зрелой miR-4513, увеличивая тем самым экспрессию гена GOSR2, продукт которого обеспечивает транспорт белков в аппарате Гольджи [97].
Полиморфизм в 3′-UTR белоккодирующих генов приводит к потере сайта связывания микроРНК в мРНК и увеличению уровня транскрипта/белка или, наоборот, к образованию нового сайта связывания микроРНК в мРНК и снижению уровня транскрипта/белка. Данные SNP представляют интерес для анализа ассоциации с заболеваниями, поскольку некоторые варианты имеют высокую популяционную частоту или существуют сильные различия в частоте SNP в различных популяциях.
В частности, ген APOC3 кодирует аполипопротеин C3, который участвует в метаболизме триглицеридов. Снижение уровня APOC3 в плазме крови приводит к снижению уровня триглицеридов и уменьшению риска развития ИБС. В 3′-UTR гена APOC3 расположен вариант rs4225:G > T. Согласно базе данных 1000 Genomes, частота аллеля T составляет 0.21 у жителей Восточной Азии, у европейцев – 0.61 и у африканцев – 0.07 [100]. В популяции Китая индивиды с генотипом TT имели более низкий уровень APOC3 и триглицеридов в плазме, этот генотип снижал также риск ИБС, поскольку аллель Т связан с супрессией трансляции APOC3 опосредованно – через взаимодействие мРНК с miR-4271 [101].
Полиморфизм в регуляторных участках генов микроРНК (промоторы, энхансеры) может приводить к нарушению уровня экcпрессии отдельных микроРНК. С использованием GWAS выявлены 69 микроРНК, расположенных в пределах 100 т.п.н. от SNP, связанных с изменением уровней липидов в крови. Функциональный анализ, проведенный на модельных животных, подтвердил связь двух микроРНК (miR-128-1 и miR-148a) с регуляцией метаболизма липидов [102].
Генетические варианты в при- и пре-микроРНК затрагивают процессинг отдельных микроРНК и, соответственно, изменяют уровень зрелой микроРНК и ее связывание с мРНК генов-мишеней. В частности, наиболее активно при атеросклерозе и его осложнениях (ИБС, инфаркт миокарда, ишемический инсульт) изучали вариант rs2910164:C>G (табл. 2). Полиморфизм rs2910164 приводит к замене G > C в последовательности пре-микроРНК гена MIR146A и далее в “пассажирской цепи” зрелой miR-146a-3p или miR-146a*. В результате нарушается процессинг и конформация вторичной структуры pre-miR-146a и ее стабильность, уменьшается продукция зрелой miR-146a в клеточной линии U2OS [103]. Показано, что генотип СС и аллель С связаны со снижением экспрессии miR-146a в клетках, что увеличивает содержание мРНК ее мишеней (IRAK1, TRAF6), внося вклад в формирование провоспалительного профиля и, следовательно, риска заболевания. Однако в тканях атеросклеротических бляшек коронарных и сонных артерий экспрессия miR-146a выше, чем в интактных сосудах [28, 34, 37]. Гены-мишени и молекулярные механизмы (пролиферация, апоптоз и миграция ГМК, модулирование иммунного ответа), посредством которых miR-146a действует в клетках артерий при их поражении атеросклерозом, активно изучаются в настоящее время [104, 105].
Вариант rs11614913:C>T (MIR196A2) также относится к SNP пре-микроРНК. Функциональный эффект этого варианта связан с нарушением процессинга pre-miR-196a2, изменением уровня зрелой miR-196a2 и последующего связывания данной микроРНК с мРНК генов-мишеней. Аллель C (генотипы СС+СT) rs11614913 ассоциирован с риском развития инфаркта миокарда у русских и осложненным течением ИБС в популяции Китая [89, 90]. Однако мета-анализ южноазиатских популяций показал, что, напротив, генотипы TT + TC rs11614913 связаны с повышенным риском развития ишемического инсульта у китайцев, но не у жителей Южной Кореи [86].
В отличие от SNP связь вариаций числа копий участков ДНК (CNV) с микроРНК и их регуляторной сетью как в норме, так и при атеросклеротическом поражении артерий практически не изучены. Сопоставление локусов микроРНК и регионов CNV выявило 289 генов микроРНК, локализованных в области CNV (miRNA-CNV) [106]. Функциональный эффект CNV в области генов микроРНК заключается в изменении их экспрессии – увеличении или уменьшении, либо в отсутствии изменений дозы генов [107]. Однако нам известна всего одна работа [108], в которой анализируется ассоциация miRNA-CNV с сердечно-сосудистыми заболеваниями, обнаружена связь CNV в генах miR-93, -122 и -192 с ИБС в сочетании с сахарным диабетом типа 2.
Ранее Nazarenko M.S. и соавт. у пяти пациентов с атеросклерозом коронарных артерий в сочетании с метаболическим синдромом выявили делецию размером примерно 39 т.п.н. в хромосомной области 3q29 (chr3:195425875-195464424; сборка генома GRCh37/hg19). Эта делеция захватывает гены MIR570 и MUC20. Согласно базе данных 1000 Genomes Consortium (Phase 3), популяционная частота CNV в этой области варьирует от 24 до 78% [109]. Более того, увеличение CNV в ней связано с повышением экспрессии miR-570 в мононуклеарах крови здоровых индивидов [110]. Более того, CNV в области miR-570 связана с риском формирования пороков развития сердца [111]. Не исключено, что эту CNV можно рассматривать в качестве кандидата для атеросклеротического поражения артерий.
Таким образом, большинство исследований ассоциации полиморфных вариантов в области генов микроРНК (miR-let-7, miR-27a, -146a, -149, -196a2, -200b, -423, -499, -618, -4513) с патологическими фенотипами атеросклероза коронарных и сонных артерий выполнены на азиатских популяциях. Наибольшее внимание привлекают полиморфизмы rs2910164:G>C (MIR146A) и rs3746444:A>G (MIR499A; MIR499B). Однако результаты этих работ противоречивы (табл. 2). Возможно, что, как и при других многофакторных заболеваниях, существенную роль в формировании предрасположенности к атеросклеротическому поражению артерий играют этнические особенности, кумулятивный эффект многих генетических вариантов, различия в коморбидности индивидов и в молекулярно-генетических механизмах формирования атеросклеротического поражения артерий разных сосудистых бассейнов.
ЗАКЛЮЧЕНИЕ
Нарушение биогенеза микроРНК, их экспрессии и регуляторной сети с генами-мишенями активно изучается, поскольку позволит идентифицировать микроРНК – селективные терапевтические мишени или биомаркеры для предсказания/прогноза развития таких клинических осложнений атеросклероза, как ИБС, инфаркт миокарда, хроническая ишемия головного мозга, ишемический инсульт [8, 112–115]. Однако вовлеченность регулома микроРНК в молекулярно-генетические механизмы формирования атеросклероза артерий разных сосудистых бассейнов малоизучена. Изучение биогенеза микроРНК, их экспрессии и регуляторной сети с генами-мишенями у человека in vivo основано на анализе одного молекулярного уровня (экспрессии микроРНК, метилирования ДНК, генетического полиморфизма).
При атеросклерозе коронарных и сонных артерий в тканях артерий и плазме/сыворотке крови изменяется экспрессия широкого спектра микроРНК (miR-let-7f, miR-1, -16, -21, -24, -29b, -106b, -133a, -145, -146a, -155, -223, -486). Эти микроРНК имеют большое количество генов-мишеней, в том числе EGFR, ABI2, IGF1R, NFAT5, BRI3BP, VEGFA, CCND1, SP1, MYC, NOTCH2, белковые продукты которых участвуют в ангиогенезе, миграции и адгезии клеток, апоптозе, активации иммунного ответа.
Метилирование ДНК может контролировать экспрессию многих генов, включая гены микроРНК. Работы, в которых анализируются изменения уровней метилирования ДНК в области генов микроРНК (miR-10b, miR-23b, локус 14q32 (miR-127, -136, -410, -431, -432 и -433), miR-143 и miR-145) в атеросклеротических бляшках, немногочисленны и проведены в цельных и разных по своей локализации сосудах. С другой стороны, подобно другим регуляторным молекулам на функцию и экспрессию микроРНК может влиять генетический полиморфизм. Ассоциацию полиморфных вариантов в области генов микроРНК (miR-let-7, miR-27a, -146a, -149, -196a2, -200b, ‑423, -499, -618, -4513) с патологическими фенотипами атеросклероза коронарных и сонных артерий изучали преимущественно с использованием отдельных SNP в азиатских популяциях и получили противоречивые результаты.
Цельное представление о молекулярных механизмах многофакторных заболеваний, включая сердечно-сосудистые, могут дать мультиомные исследования, в которых анализируют взаимодействие генетических, эпигенетических, транскриптомных, протеомных, метаболомных, экспозомных факторов [116, 117]. В частности, эпигеном тесно связан с геномом, поскольку генетические варианты в области CpG-сайтов влияют на профиль метилирования ДНК и связывание факторов транскрипции, а в области генов микроРНК – на их структуру или экспрессию. Более того, сами микроРНК могут быть мишенями для эпигенетических модификаций в том случае, когда в области промоторов генов микроРНК изменяется уровень метилирования ДНК, что приводит к изменению их экспрессии. В свою очередь, микроРНК способны регулировать уровень метилирования ДНК, воздействуя на ДНК-метилтрансферазы или метил-CpG-связанные белки. Применение такого интегративного подхода к анализу многофакторных заболеваний может дополнить существующие представления об их патогенезе, а также сгенерировать новые знания, в том числе, определить маркерные молекулы, информативные для оценки тяжести заболевания и прогноза его осложнений, идентифицировать ключевые терапевтические молекулярные мишени, обеспечивая основу для прецизионной медицины.
Исследование выполнено при финансовой поддержке Российского фонда фундаментальных исследований в рамках научного проекта № 20-115-50414.
Настоящая статья не содержит каких-либо исследований с участием людей или животных в качестве объектов исследований.
Авторы заявляют об отсутствии конфликта интересов.
Список литературы
Libby P., Buring J.E., Badimon L., Hansson G.K., Deanfield J., Bittencourt M.S., Tokgözoğlu L., Le-wis E.F. (2019) Atherosclerosis. Nat. Rev. Dis. Primers. 5(1), 56. https://doi.org/10.1038/s41572-019-0106-z
Basatemur G.L., Jørgensen H.F., Clarke M., Ben-nett M.R., Mallat Z. (2019) Vascular smooth muscle cells in atherosclerosis. Nat. Rev. Cardiol. 16(12), 727–744. https://doi.org/10.1038/s41569-019-0227-9
Chinetti-Gbaguidi G., Colin S., Staels B. (2015) Macrophage subsets in atherosclerosis. Nat. Rev. Cardiol. 12(1), 10–17. https://doi.org/10.1038/nrcardio.2014.173
Raitoharju E., Oksala N., Lehtimäki T. (2013) MicroRNAs in the atherosclerotic plaque. Clin. Chem. 59(12), 1708–1721. https://doi.org/10.1373/clinchem.2013.204917
Andreou I., Sun X., Stone P.H., Edelman E.R., Fein-berg M.W. (2015) miRNAs in atherosclerotic plaque initiation, progression, and rupture. Trends. Mol. Med. 21(5), 307–318. https://doi.org/10.1016/j.molmed.2015.02.003
Feinberg M.W., Moore K.J. (2016) MicroRNA regulation of atherosclerosis. Circ. Res. 118(4), 703–720. https://doi.org/10.1161/CIRCRESAHA.115.306300
Кучер А.Н., Назаренко М.С. (2017) Роль микро-РНК при атерогенезе. Кардиология. 57(9), 65–76.
Fasolo F., Di Gregoli K., Maegdefessel L., John-son J.L. (2019) Non-coding RNAs in cardiovascular cell biology and atherosclerosis. Cardiovasc. Res. 115(12), 1732–1756. https://doi.org/10.1093/cvr/cvz203
Friedman R.C., Farh K.K.-H., Burge C.B., Bartel D.P. (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19(1), 92–105. https://doi.org/10.1101/gr.082701.108
Catalanotto C., Cogoni C., Zardo G. (2016) MicroRNA in control of gene expression: an overview of nuclear functions. Int. J. Mol. Sci. 17(10), 1712. https://doi.org/10.3390/ijms17101712
Lu Y., Thavarajah T., Gu W., Cai J., Xu Q. (2018) Impact of miRNA in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 38(9), e159–e170. https://doi.org/10.1161/ATVBAHA.118.310227
Kozomara A., Birgaoanu M., Griffiths-Jones S. (2019) miRBase: from microRNA sequences to function. Nucl. Acids Res. 47(D1), D155–D162. https://doi.org/10.1093/nar/gky1141
Laffont B., Rayner K.J. (2017) MicroRNAs in the pathobiology and therapy of atherosclerosis. Can. J. Cardiol. 33(3), 313–324. https://doi.org/10.1016/j.cjca.2017.01.001
Olena A.F., Patton J.G. (2010) Genomic organization of microRNAs. J. Cell. Physiol. 222(3), 540–545. https://doi.org/10.1002/jcp.21993
Morales S., Monzo M., Navarro A. (2017) Epigenetic regulation mechanisms of microRNA expression. Biomol. Concepts. 8(5-6), 203–212. https://doi.org/10.1515/bmc-2017-0024
Marsico A., Huska M.R., Lasserre J., Hu H., Vuci-cevic D., Musahl A., Orom U., Vingron M. (2013) PROmiRNA: a new miRNA promoter recognition method uncovers the complex regulation of intronic miRNAs. Genome Biol. 14(8), R84. https://doi.org/10.1186/gb-2013-14-8-r84
Chakraborty C., Das S. (2016) Profiling cell-free and circulating miRNA: a clinical diagnostic tool for different cancers. Tumour Biol. 37(5), 5705–5714. https://doi.org/10.1007/s13277-016-4907-3
Wong L.L., Wang J., Liew O.W., Richards A.M., Chen Y.T. (2016) MicroRNA and heart failure. Int. J. Mol. Sci. 17(4), 502. https://doi.org/10.3390/ijms17040502
Hinske L.C., França G.S., Torres H.A., Ohara D.T., Lopes-Ramos C.M., Heyn J., Reis L.F., Ohno-Machado L., Kreth S., Galante P.A. (2014) miRIAD-integrating microRNA inter- and intragenic data. Database (Oxford). 2014, bau099. https://doi.org/10.1093/database/bau099
Huang Z., Shi J., Gao Y., Cui C., Zhang S., Li J., Zhou Y., Cui Q. (2019) HMDD v3.0: a database for experimentally supported human microRNA-disease associations. Nucl. Acids Res. 47 (D1), D1013–D1017.
Sharma H., Estep M., Birerdinc A., Afendy A., Mo-azzez A., Elariny H., Goodman Z., Chandhoke V., Baranova A., Younossi Z.M. (2013) Expression of genes for microRNA-processing enzymes is altered in advanced non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol. 28(8), 1410–1415. https://doi.org/10.1111/jgh.12268
Cipollone F., Felicioni L., Sarzani R., Ucchino S., Spigonardo F., Mandolini C., Malatesta S., Bucci M., Mammarella C., Santovito D., de Lutiis F., Marchetti A., Mezzetti A., Buttitta F. (2011) A unique microRNA signature associated with plaque instability in humans. Stroke. 42(9), 2556–2563. https://doi.org/10.1161/STROKEAHA.110.597575
Lovren F., Pan Y., Quan A., Singh K.K., Shukla P.C., Gupta N., Steer B.M., Ingram A.J., Gupta M., Al-Omran M., Teoh H., Marsden P.A., Verma S. (2012) MicroRNA-145 targeted therapy reduces atherosclerosis. Circulation. 126(11 Suppl. 1), S81–S90. https://doi.org/10.1161/CIRCULATIONAHA.111.084186
Nazari-Jahantigh M., Wei Y., Noels H., Akhtar S., Zhou Z., Koenen R.R., Heyll K., Gremse F., Kiess-ling F., Grommes J., Weber C., Schober A. (2012) MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J. Clin. Invest. 122(11), 4190–4202. https://doi.org/10.1172/JCI61716
Santovito D., Mandolini C., Marcantonio P., De Nardis V., Bucci M., Paganelli C., Magnacca F., Ucchino S., Mastroiacovo D., Desideri G., Mezzetti A., Cipollone F. (2013) Overexpression of microRNA-145 in atherosclerotic plaques from hypertensive patients. Expert. Opin. Ther. Targets. 17(3), 217–223. https://doi.org/10.1517/14728222.2013.745512
Di Gregoli K., Jenkins N., Salter R., White S., Newby A.C., Johnson J.L. (2014) MicroRNA-24 regulates macrophage behavior and retards atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 34(9), 1990–2000. https://doi.org/10.1161/ATVBAHA.114.304088
Bazan H.A., Hatfield S.A., O’Malley C.B., Brooks A.J., Lightell D. Jr., Woods T.C. (2015) Acute loss of miR-221 and miR-222 in the atherosclerotic plaque shoulder accompanies plaque rupture. Stroke. 46(11), 3285–3287. https://doi.org/10.1161/STROKEAHA.115.010567
Cao J., Zhang K., Zheng J., Dong R. (2015) MicroRNA-146a and -21 cooperate to regulate vascular smooth muscle cell proliferation via modulation of the Notch signaling pathway. Mol. Med. Rep. 11(4), 2889–2895. https://doi.org/10.3892/mmr.2014.3107
Maitrias P., Metzinger-Le Meuth V., Massy Z.A., M’Baya-Moutoula E., Reix T., Caus T., Metzinger L. (2015) MicroRNA deregulation in symptomatic carotid plaque. J. Vasc. Surg. 62(5), 1245–1250.e1. https://doi.org/10.1016/j.jvs.2015.06.136
Di Gregoli K., Mohamad Anuar N.N., Bianco R., White S.J., Newby A.C., George S.J., Johnson J.L. (2017) MicroRNA-181b controls atherosclerosis and aneurysms through regulation of TIMP-3 and elastin. Circ. Res. 120(1), 49–65. https://doi.org/10.1161/CIRCRESAHA.116.309321
Bildirici A.E., Arslan S., Özbilüm Şahin N., Berkan Ö., Beton O., Yilmaz M.B. (2018) MicroRNA-221/222 expression in atherosclerotic coronary artery plaque versus internal mammarian artery and in peripheral blood samples. Biomarkers. 23(7), 670–675. https://doi.org/10.1080/1354750X.2018.1474260
Gong F.H., Cheng W.L., Wang H., Gao M., Qin J.J., Zhang Y., Li X., Zhu X., Xia H., She Z.G. (2018) Reduced atherosclerosis lesion size, inflammatory response in miR-150 knockout mice via macrophage effects. J. Lipid. Res. 59(4), 658–669. https://doi.org/10.1194/jlr.M082651
Jin H., Li D.Y., Chernogubova E., Sun C., Busch A., Eken S.M., Saliba-Gustafsson P., Winter H., Winski G., Raaz U., Schellinger I.N., Simon N., Hegenloh R., Matic L.P., Jagodic M., Ehrenborg E., Pelisek J., Eckstein H.H., Hedin U., Backlund A., Maegdefessel L. (2018) Local delivery of miR-21 stabilizes fibrous caps in vulnerable atherosclerotic lesions. Mol. Ther. 26(4), 1040–1055. https://doi.org/10.1016/j.ymthe.2018.01.011
Raitoharju E., Lyytikäinen L.P., Levula M., Oksala N., Mennander A., Tarkka M., Klopp N., Illig T., Kähönen M., Karhunen P.J., Laaksonen R., Lehtimäki T. (2011) miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis. 219(1), 211–217. https://doi.org/10.1016/j.atherosclerosis.2011.07.020
Miller C.L., Haas U., Diaz R., Leeper N.J., Kundu R.K., Patlolla B., Assimes T.L., Kaiser F.J., Perisic L., Hedin U., Maegdefessel L., Schunkert H., Erdmann J., Quertermous T., Sczakiel G. (2014) Coronary heart disease-associated variation in TCF21 disrupts a miR-224 binding site and miRNA-mediated regulation. PLoS Genet. 10(3), e1004263. https://doi.org/10.1371/journal.pgen.1004263
Wang R., Dong L.D., Meng X.B., Shi Q., Sun W.Y. (2015) Unique microRNA signatures associated with early coronary atherosclerotic plaques. Biochem. Biophys. Res. Commun. 464(2), 574–579. https://doi.org/10.1016/j.bbrc.2015.07.010
Xue Y., Wei Z., Ding H., Wang Q., Zhou Z., Zheng S., Zhang Y., Hou D., Liu Y., Zen K., Zhang C.Y., Li J., Wang D., Jiang X. (2015) MicroRNA-19b/221/222 induces endothelial cell dysfunction via suppression of PGC-1α in the progression of atherosclerosis. Atherosclerosis. 241(2), 671–681. https://doi.org/10.1016/j.atherosclerosis.2015.06.031
Markus B., Grote K., Worsch M., Parviz B., Boening A., Schieffer B., Parahuleva M.S. (2016) Differential expression of microRNAs in endarterectomy specimens taken from patients with asymptomatic and symptomatic carotid plaques. PLoS One. 11(9), e0161632. https://doi.org/10.1371/journal.pone.0161632
Parahuleva M.S., Lipps C., Parviz B., Hölschermann H., Schieffer B., Schulz R., Euler G. (2018) MicroRNA expression profile of human advanced coronary atherosclerotic plaques. Sci. Rep. 8(1), 7823. https://doi.org/10.1038/s41598-018-25690-4
Berkan Ö., Arslan S., Lalem T., Zhang L., Şahin N.Ö., Aydemir E.I., Korkmaz Ö., Eğilmez H.R., Çekin N., Devaux Y. (2019) Regulation of microRNAs in coronary atherosclerotic plaque. Epigenomics. 11(12), 1387–1397. https://doi.org/10.2217/epi-2019-0036
Hao L., Wang X.G., Cheng J.D., You S.Z., Ma S.H., Zhong X., Quan L., Luo B. (2014) The up-regulation of endothelin-1 and down-regulation of miRNA-125a-5p, -155, and -199a/b-3p in human atherosclerotic coronary artery. Cardiovasc. Pathol. 23(4), 217–223. https://doi.org/10.1016/j.carpath.2014.03.009
Katano H., Nishikawa Y., Yamada H., Yamada K., Mase M. (2018) Differential expression of microRNAs in severely calcified carotid plaques. J. Stroke Cerebrovasc. Dis. 27(1), 108–117. https://doi.org/10.1016/j.jstrokecerebrovasdis.2017.08.009
Brennan E., Wang B., McClelland A., Mohan M., Marai M., Beuscart O., Derouiche S., Gray S., Pickering R., Tikellis C., de Gaetano M., Barry M., Belton O., Ali-Shah S.T., Guiry P., Jandeleit-Dahm K., Cooper M.E., Godson C., Kantharidis P. (2017) Protective effect of let-7 miRNA family in regulating inflammation in diabetes-associated atherosclerosis. Diabetes. 66(8), 2266–2277. https://doi.org/10.2337/db16-1405
Kern F., Aparicio-Puerta E., Li Y., Fehlmann T., Kehl T., Wagner V., Ray K., Ludwig N., Lenhof H.P., Meese E., Keller A. (2021) miRTargetLink 2.0 – interactive miRNA target gene and target pathway networks. Nucl. Acids Res. 49(W1), W409–W416. https://doi.org/10.1093/nar/gkab297
Chang L., Zhou G., Soufan O., Xia J. (2020) miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucl. Acids Res. 48(W1), W244–W251. https://doi.org/10.1093/nar/gkaa467
Kern F., Fehlmann T., Solomon J., Schwed L., Grammes N., Backes C., Van Keuren-Jensen K., Craig D.W., Meese E., Keller A. (2020) miEAA 2.0: integrating multi-species microRNA enrichment analysis and workflow management systems. Nucl. Acids Res. 48(W1), W521–W528. https://doi.org/10.1093/nar/gkaa309
Huang H.Y., Lin Y.C., Li J., Huang K.Y., Shrestha S., Hong H.C., Tang Y., Chen Y.G., Jin C.N., Yu Y., Xu J.T., Li Y.M., Cai X.X., Zhou Z.Y., Chen X.H., Pei Y.Y., Hu L., Su J.J., Cui S.D., Wang F., Xie Y.Y., Ding S.Y., Luo M.F., Chou C.H., Chang N.W., Chen K.W., Cheng Y.H., Wan X.H., Hsu W.L., Lee T.Y., Wei F.X., Huang H.D. (2020) miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucl. Acids Res. 48(D1), D148–D154. https://doi.org/10.1093/nar/gkz896
(2021) STRING: protein-protein interaction networks functional enrichment analysis https://string-db.org/
Kanuri S.H., Ipe J., Kassab K., Gao H., Liu Y., Skaar T.C., Kreutz R.P. (2018) Next generation microRNA sequencing to identify coronary artery disease patients at risk of recurrent myocardial infarction. Atherosclerosis. 278, 232–239. https://doi.org/10.1016/j.atherosclerosis.2018.09.021
Логинов В.И., Рыков С.В., Фридман М.В., Брага Э.А. (2015) Метилирование генов микроРНК и онкогенез. Биохимия. 80(2), 184–203.
Chhabra R. (2015) miRNA and methylation: a multifaceted liaison. Chembiochem. 16(2), 195–203. https://doi.org/10.1002/cbic.201402449
Ma J., Hong L., Chen Z., Nie Y., Fan D. (2014) Epigenetic regulation of microRNAs in gastric cancer. Dig. Dis. Sci. 59(4), 716–723. https://doi.org/10.1007/s10620-013-2939-8
Piletič K., Kunej T. (2016) MicroRNA epigenetic signatures in human disease. Arch. Toxicol. 90(10), 2405–2419. https://doi.org/10.1007/s00204-016-1815-7
Kunej T., Godnic I., Ferdin J., Horvat S., Dovc P., Calin G.A. (2011) Epigenetic regulation of microRNAs in cancer: an integrated review of literature. Mutat. Res. 717(1–2), 77–84. https://doi.org/10.1016/j.mrfmmm.2011.03.008
Baer C., Claus R., Plass C. (2013) Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 73(2), 473–477. https://doi.org/10.1158/0008-5472.CAN-12-3731
Wang Z., Yao H., Lin S., Zhu X., Shen Z., Lu G., Poon W.S., Xie D., Lin M.C., Kung H.F. (2013) Transcriptional and epigenetic regulation of human microRNAs. Cancer Lett. 331(1), 1–10. https://doi.org/10.1016/j.canlet.2012.12.006
Bell R.E., Golan T., Sheinboim D., Malcov H., Amar D., Salamon A., Liron T., Gelfman S., Gabet Y., Shamir R., Levy C. (2016) Enhancer methylation dynamics contribute to cancer plasticity and patient mortality. Genome Res. 26(5), 601–611. https://doi.org/10.1101/gr.197194.115
Марков А.В., Назаренко М.С., Королёва Ю.А., Лебедев И.Н., Слепцов А.А., Фролов А.В., Попов В.А., Барбараш О.Л., Барбараш Л.С., Пузырев В.П. (2014) Уровень метилирования промоторного региона гена HOXD4 у больных атеросклерозом. Медицинская генетика. 13(1), 39–42.
Wang Z., Guo D., Yang B., Wang J., Wang R., Wang X., Zhang Q. (2014) Integrated analysis of microarray data of atherosclerotic plaques: modulation of the ubiquitin-proteasome system. PLoS One. 9(10), e110288. https://doi.org/10.1371/journal.pone.0110288
Zaina S., Heyn H., Carmona F.J., Varol N., Sayols S., Condom E., Ramírez-Ruz J., Gomez A., Gonçalves I., Moran S., Esteller M. (2014) DNA methylation map of human atherosclerosis. Circ. Cardiovasc. Genet. 7(5), 692–700. https://doi.org/10.1161/CIRCGENETICS.113.000441
Aavik E., Lumivuori H., Leppänen O., Wirth T., Häkkinen S.K., Bräsen J.H., Beschorner U., Zeller T., Braspenning M., van Criekinge W., Mäkinen K., Ylä-Herttuala S. (2015) Global DNA methylation analysis of human atherosclerotic plaques reveals extensive genomic hypomethylation and reactivation at imprinted locus 14q32 involving induction of a miRNA cluster. Eur. Heart. J. 36(16), 993–1000. https://doi.org/10.1093/eurheartj/ehu437
Nazarenko M.S., Markov A.V., Lebedev I.N., Freidin M.B., Sleptcov A.A., Koroleva I.A., Frolov A.V., Popov V.A., Barbarash O.L., Puzyrev V.P. (2015) A comparison of genome-wide DNA methylation patterns between different vascular tissues from patients with coronary heart disease. PLoS One. 10(4), e0122601. https://doi.org/10.1371/journal.pone.0122601
Ehrlich K.C., Lacey M., Ehrlich M. (2019) Tissue-specific epigenetics of atherosclerosis-related ANGPT and ANGPTL genes. Epigenomics. 11(2), 169–186. https://doi.org/10.2217/epi-2018-0150
Nakahara M., Kobayashi N., Oka M., Nakano K., Okamura T., Yuo A., Saeki K. (2018) miR-10b deficiency affords atherosclerosis resistance. bioRxiv. 248641.
Shen X., Fang J., Lv X., Pei Z., Wang Y., Jiang S., Ding K. (2011) Heparin impairs angiogenesis through inhibition of microRNA-10b. J. Biol. Chem. 286(30), 26616–26627. https://doi.org/10.1074/jbc.M111.224212
Wang D., Xia M., Yan X., Li D., Wang L., Xu Y., Jin T., Ling W. (2012) Gut microbiota metabolism of anthocyanin promotes reverse cholesterol transport in mice via repressing miRNA-10b. Circ. Res. 111(8), 967–981. https://doi.org/10.1161/CIRCRESAHA.112.266502
Yu X., Li Z., Chen G., Wu W.K. (2015) MicroRNA-10b induces vascular muscle cell proliferation through Akt pathway by targeting TIP30. Curr. Vasc. Pharmacol. 13(5), 679–686. https://doi.org/10.2174/1570161113666150123112751
Goossens E.A.C., de Vries M.R., Simons K.H., Putter H., Quax P.H.A, Nossent A.Y. (2019) miRMap: profiling 14q32 microRNA expression and DNA methylation throughout the human vasculature. Front Cardiovasc. Med. 6, 113. https://doi.org/10.3389/fcvm.2019.00113
Huan T., Mendelson M., Joehanes R., Yao C., Liu C., Song C., Bhattacharya A., Rong J., Tanriverdi K., Keefe J., Murabito J.M., Courchesne P., Larson M.G., Freedman J.E., Levy D. (2020) Epigenome-wide association study of DNA methylation and microRNA expression highlights novel pathways for human complex traits. Epigenetics. 15(1–2), 183–198. https://doi.org/10.1080/15592294.2019.1640547
Edwards S.L., Beesley J., French J.D., Dunning A.M. (2013) Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 93(5), 779–797. https://doi.org/10.1016/j.ajhg.2013.10.012
Borghini A., Andreassi M.G. (2018) Genetic polymorphisms offer insight into the causal role of microRNA in coronary artery disease. Atherosclerosis. 269, 63–70. https://doi.org/10.1016/j.atherosclerosis.2017.12.022
Miao Y.R., Liu W., Zhang Q., Guo A.Y. (2018) lncRNASNP2: an updated database of functional SNPs and mutations in human and mouse lncRNAs. Nucl. Acids Res. 46(D1), D276–D280. https://doi.org/10.1093/nar/gkx1004
Moszyńska A., Gebert M., Collawn J.F., Bartoszewski R. (2017) SNPs in microRNA target miRNA sites and their potential role in human disease. Open Biol. 7(4), 170019. https://doi.org/10.1098/rsob.170019
Zhang L., Yang J., Xue Q., Yang D., Lu Y., Guang X., Zhang W., Ba R., Zhu H., Ma X. (2016) An rs13293512 polymorphism in the promoter of let-7 is associated with a reduced risk of ischemic stroke. J. Thromb. Thrombolysis. 42(4), 610–615. https://doi.org/10.1007/s11239-016-1400-1
Cai M.Y., Cheng J., Zhou M.Y., Liang L.L., Lian S.M., Xie X.S., Xu S., Liu X., Xiong X.D. (2018) The association between pre-miR-27a rs895819 polymorphism and myocardial infarction risk in a Chinese han population. Lipids Health Dis. 17(1), 7. https://doi.org/10.1186/s12944-017-0652-x
Oner T., Arslan C., Yenmis G., Arapi B., Tel C., Ayde-mir B., Sultuybek G.K. (2017) Association of NFKB1A and microRNAs variations and the susceptibility to atherosclerosis. J. Genet. 96(2), 251–259. https://doi.org/10.1007/s12041-017-0768-9
He Y., Yang J., Kong D., Lin J., Xu C., Ren H., Ouyang P., Ding Y., Wang K. (2015) Association of miR-146a rs2910164 polymorphism with cardio-cerebrovascular diseases: a systematic review and meta-analysis. Gene. 565(2), 171–179. https://doi.org/10.1016/j.gene.2015.04.020
Xiong X.D., Cho M., Cai X.P., Cheng J., Jing X., Cen J.M., Liu X., Yang X.L., Suh Y. (2014) A common variant in pre-miR-146 is associated with coronary artery disease risk and its mature miRNA expression. Mutat. Res. 761, 15–20. https://doi.org/10.1016/j.mrfmmm.2014.01.001
Bastami M., Choupani J., Saadatian Z., Zununi Vahed S., Mansoori Y., Daraei A., Samadi Kafil H., Masotti A., Nariman-Saleh-Fam Z. (2019) Polymorphisms and risk of cardio-cerebrovascular diseases: a systematic review and meta-analysis. Int. J. Mol. Sci. 20(2), 293. https://doi.org/10.3390/ijms20020293
Bao M.H., Xiao Y., Zhang Q.S., Luo H.Q., Luo J., Zhao J., Li G.Y., Zeng J., Li J.M. (2015) Meta-analysis of miR-146a polymorphisms association with coronary artery diseases and ischemic stroke. Int. J. Mol. Sci. 16(7), 14305–14317. https://doi.org/10.3390/ijms160714305
Hamann L., Glaeser C., Schulz S., Gross M., Franke A., Nöthlings U., Schumann R.R. (2014) A micro RNA-146a polymorphism is associated with coronary restenosis. Int. Immunogenet. 41(5), 393–396. https://doi.org/10.1111/iji.12136
Shen J., Zhang M., Sun M., Tang K., Zhou B. (2015) The relationship of miR-146a gene polymorphism with carotid atherosclerosis in Chinese patients with type 2 diabetes mellitus. Thromb. Res. 136(6), 1149–1155. https://doi.org/10.1016/j.thromres.2015.10.013
Zhu R., Liu X., He Z., Li Q. (2014) miR-146a and miR-196a2 polymorphisms in patients with ischemic stroke in the northern Chinese han population. Neurochem. Res. 39(9), 1709–1716. https://doi.org/10.1007/s11064-014-1364-5
Zhong H., Cai Y., Cheng J., Cai D., Chen L., Su C., Li K., Chen P., Xu J., Cui L. (2016) Apolipoprotein E epsilon 4 enhances the association between the rs2910164 polymorphism of miR-146a and risk of atherosclerotic cerebral infarction. J. Atheroscler. Thromb. 23(7), 819–829. https://doi.org/10.5551/jat.32904
Jeon Y.J., Kim O.J., Kim S.Y., Oh S.H., Oh D., Kim O.J., Shin B.S., Kim N.K. (2013). Association of the miR-146a, miR-149, miR-196a2, and miR-499 polymorphisms with ischemic stroke and silent brain infarction risk. Arterioscler. Thromb. Vasc. Biol. 33(2), 420–430. https://doi.org/10.1161/ATVBAHA.112.300251
Zhu J., Yue H., Qiao C., Li Y. (2015) Association between single-nucleotide polymorphism (SNP) in miR-146a, miR-196a2, and miR-499 and risk of ischemic stroke: a meta-analysis. Med. Sci. Monit. 21, 3658–3663. https://doi.org/10.12659/msm.895233
Qu J.Y., Xi J., Zhang Y.H., Zhang C.N., Song L., Song Y., Hui R.T., Chen J.Z. (2016) Association of the microRNA-146a SNP rs2910164 with ischemic stroke incidence and prognosis in a Chinese population. Int. J. Mol. Sci. 17(5), 660. https://doi.org/10.3390/ijms17050660
Sung J.H., Kim S.H., Yang W.I., Kim W.J., Moon J.Y., Kim I.J., Cha D.H., Cho S.Y., Kim J.O., Kim K.A., Kim O.J., Lim S.W., Kim N.K. (2016) miRNA polymorphisms (miR‑146a, miR‑149, miR‑196a2 and miR‑499) are associated with the risk of coronary artery disease. Mol. Med. Rep. 14(3), 2328–2342. https://doi.org/10.3892/mmr.2016.5495
Zhi H., Wang L., Ma G., Ye X., Yu X., Zhu Y., Zhang Y., Zhang J., Wang B. (2012) Polymorphisms of miRNAs genes are associated with the risk and prognosis of coronary artery disease. Clin. Res. Cardiol. 101(4), 289–296.https://doi.org/10.1007/s00392-011-0391-3
Осьмак Г.Ж., Матвеева Н.А., Титов Б.В., Фаворова О.О. (2018) Связь полиморфизма гена MIR196A2 с инфарктом миокарда и возможное вовлечение микроРНК miR-196a2 в сигнальные пути, участвующие в формировании патологического фенотипа. Молекуляр. биология. 52(6), 1006–1013.
Kim J., Choi G.H., Ko K.H., Kim J.O., Oh S.H., Park Y.S., Kim O.J., Kim N.K. (2016) Association of the single nucleotide polymorphisms in microRNAs 130b, 200b, and 495 with ischemic stroke susceptibility and post-stroke mortality. PLoS One. 11(9), e0162519. https://doi.org/10.1371/journal.pone.0162519
Jha C.K., Mir R., Elfaki I., Khullar N., Rehman S., Javid J., Banu S., Chahal S. (2019) Potential impact of microRNA-423 gene variability in coronary artery disease. Endocr. Metab. Immune Disord. Drug Targets. 19(1), 67–74. https://doi.org/10.2174/1871530318666181005095724
Chen C., Hong H., Chen L., Shi X., Chen Y., Weng Q. (2014) Association of microRNA polymorphisms with the risk of myocardial infarction in a Chinese population. Tohoku J. Exp. Med. 233(2), 89–94. https://doi.org/10.1620/tjem.233.89
Darabi H., Salmaninejad A., Jaripour M.E., Azarpaz-hooh M.R., Mojarrad M., Sadr-Nabavi A. (2019) Association of the genetic polymorphisms in immunoinflammatory microRNAs with risk of ischemic stroke and subtypes in an Iranian population. J. Cell. Physiol. 234(4), 3874–3886. https://doi.org/10.1002/jcp.27159
Liu Y., Ma Y., Zhang B., Wang S.X., Wang X.M., Yu J.M. (2014) Genetic polymorphisms in pre-microRNAs and risk of ischemic stroke in a Chinese population. J. Mol. Neurosci. 52(4), 473–480. https://doi.org/10.1007/s12031-013-0152-z
Zhang Z., Xu G., Cai B., Zhang H., Zhu W., Liu X. (2017) Genetic variants in microRNAs predict recurrence of ischemic stroke. Mol. Neurobiol. 54(4), 2776–2780. https://doi.org/10.1007/s12035-016-9865-7
Ghanbari M., de Vries P.S., de Looper H., Peters M.J., Schurmann C., Yaghootkar H., Dörr M., Frayling T.M., Uitterlinden A.G., Hofman A., van Meurs J.B., Erke-land S.J., Franco O.H., Dehghan A. (2014) A genetic variant in the seed region of miR-4513 shows pleiotropic effects on lipid and glucose homeostasis, blood pressure, and coronary artery disease. Hum. Mutat. 35(12), 1524–1531. https://doi.org/10.1002/humu.22706
(2021) Reference SNP (rs) Report: rs2910164. https://www.ncbi.nlm.nih.gov/snp/rs2910164
Li Q., Chen L., Chen D., Wu X., Chen M. (2015) Influence of microRNA-related polymorphisms on clinical outcomes in coronary artery disease. Am. J. Transl. Res. 7(2), 393–400.
(2021) Ensembl rs4225 SNP Allele: frequency (count) https://www.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=11:116832455-116833455;v=rs4225;vdb=variation;vf=164407333
Hu S.L., Cui G.L., Huang J., Jiang J.G., Wang D.W. (2016) An APOC3 3'UTR variant associated with plasma triglycerides levels and coronary heart disease by creating a functional miR-4271 binding site. Sci. Rep. 6, 32700. https://doi.org/10.1038/srep32700
Wagschal A., Najafi-Shoushtari S.H., Wang L., Goedeke L., Sinha S., deLemos A.S., Black J.C., Ra-mírez C.M., Li Y., Tewhey R., Hatoum I., Shah N., Lu Y., Kristo F., Psychogios N., Vrbanac V., Lu Y.C., Hla T., de Cabo R., Tsang J.S., Schadt E., Sabeti P.C., Kathiresan S., Cohen D.E., Whetstine J., Chung R.T., Fernández-Hernando C., Kaplan L.M., Bernards A., Gerszten R.E., Näär A.M. (2015) Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat. Med. 21(11), 1290–1297. https://doi.org/10.1038/nm.3980
Jazdzewski K., Murray E.L., Franssila K., Jarzab B., Schoenberg D.R., de la Chapelle A. (2008) Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA. 105(20), 7269–7274. https://doi.org/10.1073/pnas.0802682105
Wang D., Atanasov A.G. (2019) The microRNAs regulating vascular smooth muscle cell proliferation: a minireview. Int. J. Mol. Sci. 20(2), 324. https://doi.org/10.3390/ijms20020324
Sun X., Icli B., Wara A.K., Belkin N., He S., Kobzik L., Hunninghake G.M., Vera M.P., MICU Registry, Blackwell T.S., Baron R.M., Feinberg M.W. (2012) MicroRNA-181b regulates NF-κB-mediated vascular inflammation. J. Clin. Invest. 122(6), 1973–1990. https://doi.org/10.1172/JCI61495
Dweep H., Georgiou G.D., Gretz N., Deltas C., Voska-rides K., Felekkis K. (2013) CNVs-microRNAs interactions demonstrate unique characteristics in the human genome. An interspecies in silico analysis. PLoS One. 8(12), e81204. https://doi.org/10.1371/journal.pone.0081204
Marcinkowska M., Szymanski M., Krzyzosiak W.J., Kozlowski P. (2011) Copy number variation of microRNA genes in the human genome. BMC Genomics. 12, 183. https://doi.org/10.1186/1471-2164-12-183
Sohrabifar N., Ghaderian S., Vakili H., Ghaedi H., Rouhani B., Jafari H., Heidari L. (2021) MicroRNA-copy number variations in coronary artery disease patients with or without type 2 diabetes mellitus. Arch. Physiol. Biochem. 127(6), 497–503. https://doi.org/10.1080/13813455.2019.1651340
Nazarenko M.S., Sleptcov A.A., Lebedev I.N., Skrya-bin N.A., Markov A.V., Golubenko M.V., Koroleva I.A., Kazancev A.N., Barbarash O.L., Puzyrev V.P. (2017) Genomic structural variations for cardiovascular and metabolic comorbidity. Sci. Rep. 7, 41268. https://doi.org/10.1038/srep41268
Lins T.C. de L. (2014) Variação estrutural no número de cópias e sua implicação na expressão de microRNA em humanos. https://repositorio.unb.br/handle/10482/16506
Xing H.J., Li Y.J., Ma Q.M., Wang A.M., Wang J.L., Sun M., Jian Q., Hu J.H., Li D., Wang L. (2013) Identification of microRNAs present in congenital heart disease associated copy number variants. Eur. Rev. Med. Pharmacol Sci. 17(15), 2114–2120.
Chen L.J., Lim S.H., Yeh Y.T., Lien S.C., Chiu J.J. (2012) Roles of microRNAs in atherosclerosis and restenosis. J. Biomed. Sci. 19(1), 79. https://doi.org/10.1186/1423-0127-19-79
Schober A., Weber C. (2016) Mechanisms of microRNAs in atherosclerosis. Annu. Rev. Pathol. 11, 583–616. https://doi.org/10.1146/annurev-pathol-012615-044135
Johnson J.L. (2019) Elucidating the contributory role of microRNA to cardiovascular diseases (a review). Vascul. Pharmacol. 114, 31–48. https://doi.org/10.1016/j.vph.2018.10.010
Tao J., Xia L., Cai Z., Liang L., Chen Y., Meng J., Wang Z. (2021) Interaction between microRNA and DNA methylation in atherosclerosis. DNA Cell Biol. 40(1), 101–115. https://doi.org/10.1089/dna.2020.6138
Mens M., Maas S., Klap J., Weverling G.J., Klat-ser P., Brakenhoff J., van Meurs J., Uitterlinden A.G., Ikram M.A., Kavousi M., Ghanbari M. (2020) Multi-omics analysis reveals microRNAs associated with cardiometabolic traits. Front Genet. 11, 110. https://doi.org/10.3389/fgene.2020.00110
Vohra M., Sharma A.R., Prabhu B.N., Rai P.S. (2020) SNPs in sites for DNA methylation, transcription factor binding, and miRNA targets leading to allele-specific gene expression and contributing to complex disease risk: a systematic review. Public Health Genomics. 23(5–6), 155–170. https://doi.org/10.1159/000510253
Дополнительные материалы отсутствуют.
Инструменты
Молекулярная биология


